Abstract
Background
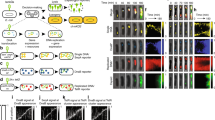
The CRISPR-Cas system is a widespread prokaryotic defense system which targets and cleaves invasive nucleic acids, such as plasmids or viruses. So far, a great number of studies have focused on the components and mechanisms of this system, however, a direct visualization of CRISPR-Cas degrading invading DNA in real-time has not yet been studied at the single-cell level.
Methods
In this study, we fluorescently label phage lambda DNA in vivo, and track the labeled DNA over time to characterize DNA degradation at the single-cell level.
Results
At the bulk level, the lysogenization frequency of cells harboring CRISPR plasmids decreases significantly compared to cells with a non-CRISPR control. At the single-cell level, host cells with CRISPR activity are unperturbed by phage infection, maintaining normal growth like uninfected cells, where the efficiency of our anti-lambda CRISPR system is around 26%. During the course of time-lapse movies, the average fluorescence of invasive phage DNA in cells with CRISPR activity, decays more rapidly compared to cells without, and phage DNA is fully degraded by around 44 minutes on average. Moreover, the degradation appears to be independent of cell size or the phage DNA ejection site suggesting that Cas proteins are dispersed in sufficient quantities throughout the cell.
Conclusions
With the CRISPR-Cas visualization system we developed, we are able to examine and characterize how a CRISPR system degrades invading phage DNA at the single-cell level. This work provides direct evidence and improves the current understanding on how CRISPR breaks down invading DNA.

Article PDF
Similar content being viewed by others
References
Mohanraju, P., Makarova, K. S., Zetsche, B., Zhang, F., Koonin, E. V. and van der Oost, J. (2016) Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science, 353, aad5147
Deveau, H., Garneau, J. E. and Moineau, S. (2010) CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol., 64, 475–493
van Erp, P. B., Jackson, R. N., Carter, J., Golden, S. M., Bailey, S. and Wiedenheft, B. (2015) Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli. Nucleic Acids Res., 43, 8381–8391
Bhaya, D., Davison, M. and Barrangou, R. (2011) CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet., 45, 273–297
Sternberg, S. H., Richter, H., Charpentier, E. and Qimron, U. (2016) Adaptation in CRISPR-Cas systems. Mol. Cell, 61, 797–808
van der Oost, J., Westra, E. R., Jackson, R. N. and Wiedenheft, B. (2014) Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol., 12, 479–492
Huo, Y., Nam, K. H., Ding, F., Lee, H., Wu, L., Xiao, Y., Farchione, M. D. Jr, Zhou, S., Rajashankar, K., Kurinov, I., et al. (2014) Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat. Struct. Mol. Biol., 21, 771–777
Hatoum-Aslan, A., Maniv, I. and Marraffini, L. A. (2011) Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc. Natl. Acad. Sci. USA, 108, 21218–21222
Sinkunas, T., Gasiunas, G., Fremaux, C., Barrangou, R., Horvath, P. and Siksnys, V. (2011) Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J., 30, 1335–1342
Barrangou, R. (2015) Diversity of CRISPR-Cas immune systems and molecular machines. Genome Biol., 16, 247
Mulepati, S. and Bailey, S. (2013) In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J. Biol. Chem., 288, 22184–22192
Amitai, G. and Sorek, R. (2016) CRISPR-Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol., 14, 67–76
Künne, T., Kieper, S. N., Bannenberg, J.W., Vogel, A. I., Miellet, W. R., Klein, M., Depken, M., Suarez-Diez, M. and Brouns, S. J. (2016) Cas3-derived target DNA degradation fragments fuel primed CRISPR adaptation. Mol. Cell, 63, 852–864
McGinn, J. and Marraffini, L. A. (2016) CRISPR-Cas systems optimize their immune response by specifying the site of spacer integration. Mol. Cell, 64, 616–623
Jackson, R. N., Golden, S. M., van Erp, P. B., Carter, J., Westra, E. R., Brouns, S. J., van der Oost, J., Terwilliger, T. C., Read, R. J. and Wiedenheft, B. (2014) Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science, 345, 1473–1479
Hochstrasser, M. L., Taylor, D. W., Bhat, P., Guegler, C. K., Sternberg, S. H., Nogales, E. and Doudna, J. A. (2014) CasA mediates Cas3- catalyzed target degradation during CRISPR RNA-guided interference. Proc. Natl. Acad. Sci. USA, 111, 6618–6623
Westra, E. R., van Erp, P. B., Kunne, T., Wong, S. P., Staals, R. H., Seegers, C. L., Bollen, S., Jore, M. M., Semenova, E., Severinov, K., et al. (2012) CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell, 46, 595–605
Mulepati, S., Heroux, A. and Bailey, S. (2014) Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science, 345, 1479–1484
Redding, S., Sternberg, S. H., Marshall, M., Gibb, B., Bhat, P., Guegler, C. K., Wiedenheft, B., Doudna, J. A. and Greene, E. C. (2015) Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas system. Cell, 163, 854–865
Brouns, S. J., Jore, M. M., Lundgren, M., Westra, E. R., Slijkhuis, R. J., Snijders, A. P., Dickman, M. J., Makarova, K. S., Koonin, E. V. and van der Oost, J. (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science, 321, 960–964
Edgar, R. and Qimron, U. (2010) The Escherichia coli CRISPR system protects from lambda lysogenization, lysogens, and prophage induction. J. Bacteriol., 192, 6291–6294
Babic, A., Lindner, A. B., Vulic, M., Stewart, E. J. and Radman, M. (2008) Direct visualization of horizontal gene transfer. Science, 319, 1533–1536
Shao, Q., Hawkins, A. and Zeng, L. (2015) Phage DNA dynamics in cells with different fates. Biophys. J., 108, 2048–2060
Pul, U., Wurm, R., Arslan, Z., Geissen, R., Hofmann, N. and Wagner, R. (2010) Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol., 75, 1495–1512
Lu, M., Campbell, J. L., Boye, E. and Kleckner, N. (1994) SeqA: a negative modulator of replication initiation in E. coli. Cell, 77, 413–426
Slater, S., Wold, S., Lu, M., Boye, E., Skarstad, K. and Kleckner, N. (1995) E. coli SeqA protein binds oriC in two different methylmodulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell, 82, 927–936
Pougach, K., Semenova, E., Bogdanova, E., Datsenko, K. A., Djordjevic, M., Wanner, B. L. and Severinov, K. (2010) Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol., 77, 1367–1379
Shao, Q., Trinh, J. T., McIntosh, C. S., Christenson, B., Balazsi, G. and Zeng, L. (2017) Lysis-lysogeny coexistence: prophage integration during lytic development. Microbiology Open, 6
Van Valen, D., Wu, D., Chen, Y. J., Tuson, H., Wiggins, P. and Phillips, R. (2012) A single-molecule Hershey-Chase experiment. Curr. Biol., 22, 1339–1343
Edgar, R., Rokney, A., Feeney, M., Semsey, S., Kessel, M., Goldberg, M. B., Adhya, S. and Oppenheim, A. B. (2008) Bacteriophage infection is targeted to cellular poles. Mol. Microbiol., 68, 1107–1116
Rothenberg, E., Sepulveda, L. A., Skinner, S. O., Zeng, L., Selvin, P. R. and Golding, I. (2011) Single-virus tracking reveals a spatial receptor-dependent search mechanism. Biophys. J., 100, 2875–2882
Zeng, L., Skinner, S. O., Zong, C., Sippy, J., Feiss, M. and Golding, I. (2010) Decision making at a subcellular level determines the outcome of bacteriophage infection. Cell, 141, 682–691
Zeng, L. and Golding, I. (2011) Following cell-fate in E. coli after infection by phage lambda. J. Vis. Exp., 56, e3363
Sliusarenko, O., Heinritz, J., Emonet, T. and Jacobs-Wagner, C. (2011) High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol., 80, 612–627
Acknowledgements
We are grateful to Rodem Edgar for providing the CRISPR plasmids. We would like to thank all members of the Zeng laboratory for help with the experiments and data analysis. Work in the Zeng laboratory was supported by the National Institutes of Health (R01GM107597). The funder had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is dedicated to the Special Collection of Synthetic Biology, Aiming for Quantitative Control of Cellular Systems (Eds. Cheemeng Tan and Haiyan Liu).
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Guan, J., Shi, X., Burgos, R. et al. Visualization of phage DNA degradation by a type I CRISPR-Cas system at the single-cell level. Quant Biol 5, 67–75 (2017). https://doi.org/10.1007/s40484-017-0099-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40484-017-0099-0