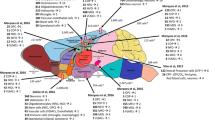
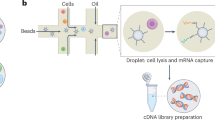
Abstract
Purpose of Review
This review summarizes some of the most recent studies on single-cell resolution sequencing of the post-mortem human brain and the application of these techniques for the study of psychiatric and neurological disorders.
Recent Findings
Over the last several years, researchers have optimized single-cell transcriptome and genome sequencing in post-mortem human brain tissue. This has given us unprecedented access to cell-type specific gene expression profiles and somatic mutations unique to pathological states. Additionally, we can now measure epigenetic information from individual brain cells and with advanced statistical approaches, we can focus on the key cell-types underlying psychiatric and other brain phenotypes.
Summary
A new era of cell-type specific and single-cell resolution studies of the human brain is underway. With proper application of rapidly advancing laboratory and data analysis techniques, we should witness important advances in our understanding of molecular changes associated with psychiatric and neurological phenotypes.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Stuart T, Satija R. Integrative single-cell analysis. Nat Rev Genet. 2019;20:257–72. https://doi.org/10.1038/s41576-019-0093-7.
Kulkarni A, Anderson AG, Merullo DP, Konopka G. Beyond bulk: a review of single cell transcriptomics methodologies and applications. Curr Opin Biotechnol. 2019;58:129–36. https://doi.org/10.1016/j.copbio.2019.03.001.
Krishnaswami SR, Grindberg RV, Novotny M, Venepally P, Lacar B, Bhutani K, et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc. 2016;11(3):499–524. https://doi.org/10.1038/nprot.2016.015.
Johnson MB, Wang PP, Atabay KD, Murphy EA, Doan RN, Hecht JL, et al. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat Neurosci. 2015;18:637–46. https://doi.org/10.1038/nn.3980. https://www.nature.com/articles/nn.3980#supplementary-information.
Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci. 2015;112(23):7285–90. https://doi.org/10.1073/pnas.1507125112.
Macosko Evan Z, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using Nanoliter droplets. Cell. 2015;161(5):1202–14. https://doi.org/10.1016/j.cell.2015.05.002.
Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM, et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc. 2016;12:44–73. https://doi.org/10.1038/nprot.2016.154. https://www.nature.com/articles/nprot.2016.154#supplementary-information.
Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–8. https://doi.org/10.1038/nmeth.4407. https://www.nature.com/articles/nmeth.4407#supplementary-information.
Hu P, Fabyanic E, Kwon DY, Tang S, Zhou Z, Wu H. Dissecting cell-type composition and activity-dependent transcriptional state in mammalian brains by massively parallel single-nucleus RNA-Seq. Mol Cell. 2017;68(5):1006–15.e7. https://doi.org/10.1016/j.molcel.2017.11.017.
Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R et al. Massively parallel digital transcriptional profiling of single cells. 2017;8:14049. doi:https://doi.org/10.1038/ncomms14049. https://www.nature.com/articles/ncomms14049#supplementary-information,
Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357(6352):661–7. https://doi.org/10.1126/science.aam8940.
Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360(6385):176–82. https://doi.org/10.1126/science.aam8999.
Mulqueen RM, Pokholok D, Norberg SJ, Torkenczy KA, Fields AJ, Sun D, et al. Highly scalable generation of DNA methylation profiles in single cells. Nat Biotechnol. 2018;36:428–31. https://doi.org/10.1038/nbt.4112. https://www.nature.com/articles/nbt.4112#supplementary-information.
Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell. 2018;174(5):1309–24.e18. https://doi.org/10.1016/j.cell.2018.06.052.
Preissl S, Fang R, Huang H, Zhao Y, Raviram R, Gorkin DU, et al. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat Neurosci. 2018;21(3):432–9. https://doi.org/10.1038/s41593-018-0079-3.
Sinnamon JR, Torkenczy KA, Linhoff MW, Vitak SA, Mulqueen RM, Pliner HA, et al. The accessible chromatin landscape of the murine hippocampus at single-cell resolution. Genome Res. 2019;29(5):857–69. https://doi.org/10.1101/gr.243725.118.
Cao J, Cusanovich DA, Ramani V, Aghamirzaie D, Pliner HA, Hill AJ, et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361(6409):1380–5. https://doi.org/10.1126/science.aau0730.
Pellegrino M, Sciambi A, Treusch S, Durruthy-Durruthy R, Gokhale K, Jacob J, et al. High-throughput single-cell DNA sequencing of acute myeloid leukemia tumors with droplet microfluidics. Genome Res. 2018;28(9):1345–52. https://doi.org/10.1101/gr.232272.117.
Lareau CA, Duarte FM, Chew JG, Kartha VK, Burkett ZD, Kohlway AS, et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat Biotechnol. 2019;37:916–24. https://doi.org/10.1038/s41587-019-0147-6.
Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–8. https://doi.org/10.1038/nmeth.4380. https://www.nature.com/articles/nmeth.4380#supplementary-information.
Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, et al. Electrophysiological, transcriptomic and morphologic profiling of single neurons using patch-seq. Nat Biotechnol. 2015;34:199–203. https://doi.org/10.1038/nbt.3445. https://www.nature.com/articles/nbt.3445#supplementary-information.
Lake BB, Codeluppi S, Yung YC, Gao D, Chun J, Kharchenko PV, et al. A comparative strategy for single-nucleus and single-cell transcriptomes confirms accuracy in predicted cell-type expression from nuclear RNA. Sci Rep. 2017;7(1):6031. https://doi.org/10.1038/s41598-017-04426-w.
Grindberg RV, Yee-Greenbaum JL, McConnell MJ, Novotny M, O’Shaughnessy AL, Lambert GM, et al. RNA-sequencing from single nuclei. Proc Natl Acad Sci. 2013;110(49):19802–7. https://doi.org/10.1073/pnas.1319700110.
Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–42. https://doi.org/10.1126/science.aaa1934.
Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–46. https://doi.org/10.1038/nn.4216. https://www.nature.com/articles/nn.4216#supplementary-information.
Hrvatin S, Hochbaum DR, Nagy MA, Cicconet M, Robertson K, Cheadle L, et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci. 2018;21(1):120–9. https://doi.org/10.1038/s41593-017-0029-5.
Kalish BT, Cheadle L, Hrvatin S, Nagy MA, Rivera S, Crow M, et al. Single-cell transcriptomics of the developing lateral geniculate nucleus reveals insights into circuit assembly and refinement. Proc Natl Acad Sci. 2018;115(5):E1051–60. https://doi.org/10.1073/pnas.1717871115.
Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, et al. Div-Seq: single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353(6302):925–8. https://doi.org/10.1126/science.aad7038.
Arneson D, Zhang G, Ying Z, Zhuang Y, Byun HR, Ahn IS, et al. Single cell molecular alterations reveal target cells and pathways of concussive brain injury. Nat Commun. 2018;9(1):3894. https://doi.org/10.1038/s41467-018-06222-0.
Hochgerner H, Zeisel A, Lönnerberg P, Linnarsson S. Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat Neurosci. 2018;21(2):290–9. https://doi.org/10.1038/s41593-017-0056-2.
Tiklová K, Björklund ÅK, Lahti L, Fiorenzano A, Nolbrant S, Gillberg L, et al. Single-cell RNA sequencing reveals midbrain dopamine neuron diversity emerging during mouse brain development. Nat Commun. 2019;10(1):581. https://doi.org/10.1038/s41467-019-08453-1.
La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell. 2016;167(2):566–80.e19. https://doi.org/10.1016/j.cell.2016.09.027.
Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcao A, Xiao L, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352(6291):1326–9. https://doi.org/10.1126/science.aaf6463.
Marques S, Vanichkina D, van Bruggen D, Floriddia E, Munguba H, Varemo L et al. Single-cell transcriptomic profiling of progenitors of the oligodendrocyte lineage reveals transcriptional convergence during development. bioRxiv. 2017.
Mathys H, Adaikkan C, Gao F, Young JZ, Manet E, Hemberg M, et al. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 2017;21(2):366–80. https://doi.org/10.1016/j.celrep.2017.09.039.
Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50(1):253–71.e6. https://doi.org/10.1016/j.immuni.2018.11.004.
Sousa C, Golebiewska A, Poovathingal SK, Kaoma T, Pires-Afonso Y, Martina S, et al. Single-cell transcriptomics reveals distinct inflammation-induced microglia signatures. EMBO Rep. 2018;19(11):e46171. https://doi.org/10.15252/embr.201846171.
Lacar B, Linker SB, Jaeger BN, Krishnaswami SR, Barron JJ, Kelder MJE, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7:11022. https://doi.org/10.1038/ncomms11022 https://www.nature.com/articles/ncomms11022#supplementary-information.
Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, et al. Molecular architecture of the mouse nervous system. Cell. 2018;174(4):999–1014.e22. https://doi.org/10.1016/j.cell.2018.06.021.
Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell. 2018;174(4):1015–30.e16. https://doi.org/10.1016/j.cell.2018.07.028.
Häring M, Zeisel A, Hochgerner H, Rinwa P, Jakobsson JET, Lönnerberg P, et al. Neuronal atlas of the dorsal horn defines its architecture and links sensory input to transcriptional cell types. Nat Neurosci. 2018;21(6):869–80. https://doi.org/10.1038/s41593-018-0141-1.
Cuevas-Diaz Duran R, Wei H, Wu JQ. Single-cell RNA-sequencing of the brain. Clin Transl Med. 2017;6(1):20. https://doi.org/10.1186/s40169-017-0150-9.
Ofengeim D, Giagtzoglou N, Huh D, Zou C, Yuan J. Single-cell RNA sequencing: unraveling the brain one cell at a time. Trends Mol Med. 2017;23(6):563–76. https://doi.org/10.1016/j.molmed.2017.04.006.
Guillaumet-Adkins A, Heyn H. Single-cell genomics unravels brain cell-type complexity. In: Delgado-Morales R, editor. Neuroepigenomics in aging and disease. Cham: Springer International Publishing; 2017. p. 393–407.
Cembrowski MS. Single-cell transcriptomics as a framework and roadmap for understanding the brain. J Neurosci Methods. 2019;326:108353. https://doi.org/10.1016/j.jneumeth.2019.108353.
Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352(6293):1586–90. https://doi.org/10.1126/science.aaf1204.
•• Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol. 2018;36(1):70–80. https://doi.org/10.1038/nbt.4038This study measured both gene expression and chromatin accessibility in individual cells at high-throughput in mulitple regions of the post-mortem human brain. Single-nucleus gene expression was used to refine cell-type classfication based on single-nucleus chromatin accessibility, and cell-type specific chromatin accessibility was used to assess cell-type contributions to neurological and psychiatric diseases.
•• Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ. Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell. 2019;177(7):1873–87.e17. https://doi.org/10.1016/j.cell.2019.05.006This study introduces LIGER, a computational tool for analysis of single-cell sequencing data which allows aligment of multiple datasets into a single consolidated dataset while preserving dataset specific information and allows for integration of multi-modal data.
Boldog E, Bakken TE, Hodge RD, Novotny M, Aevermann BD, Baka J, et al. Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat Neurosci. 2018;21(9):1185–95. https://doi.org/10.1038/s41593-018-0205-2.
Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, et al. Conserved cell types with divergent features between human and mouse cortex. bioRxiv. 2018:384826. https://doi.org/10.1101/384826.
•• Renthal W, Boxer LD, Hrvatin S, Li E, Silberfeld A, Nagy MA, et al. Characterization of human mosaic Rett syndrome brain tissue by single-nucleus RNA sequencing. Nat Neurosci. 2018;21(12):1670–9. https://doi.org/10.1038/s41593-018-0270-6This study demonstrates the possibility of studying somatic mutations using high-throughput 3'-biased single-nucleus RNA-seq and the technique is widely applicable for studying cell-type specific contributions to X-linked neurological conditions.
• Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, et al. Single-cell genomics identifies cell type–specific molecular changes in autism. Science. 2019;364(6441):685. https://doi.org/10.1126/science.aav8130This study reports for the first time the cell-type specific contributions to autism spectrum disorders in the prefrontal and anterior cingulate cortex at single-cell resolution.
Sorrells SF, Paredes MF, Velmeshev D, Herranz-Pérez V, Sandoval K, Mayer S, et al. Immature excitatory neurons develop during adolescence in the human amygdala. Nat Commun. 2019;10(1):2748. https://doi.org/10.1038/s41467-019-10765-1.
• Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570(7761):332–7. https://doi.org/10.1038/s41586-019-1195-2This study reports for the first time the cell-type specific contributions to Alzheimer's disease in the human prefrontal cortex at single-cell resolution.
Jäkel S, Agirre E, Mendanha Falcão A, van Bruggen D, Lee KW, Knuesel I, et al. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature. 2019;566(7745):543–7. https://doi.org/10.1038/s41586-019-0903-2.
Nagy C, Maitra M, Suderman M, Theroux J-F, Mechawar N, Ragoussis J, et al. Single-nucleus RNA sequencing shows convergent evidence from different cell types for altered synaptic plasticity in major depressive disorder. bioRxiv. 2018. https://doi.org/10.1101/384479.
Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA. Religious orders study and rush memory and aging project. J Alzheimer's Dis : JAD. 2018;64(s1):S161–s89. https://doi.org/10.3233/jad-179939.
Singer T, McConnell MJ, Marchetto MCN, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33(8):345–54. https://doi.org/10.1016/j.tins.2010.04.001.
McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, et al. Mosaic copy number variation in human neurons. Science. 2013;342(6158):632–7. https://doi.org/10.1126/science.1243472.
Cai X, Evrony Gilad D, Lehmann Hillel S, Elhosary Princess C, Mehta Bhaven K, Poduri A, et al. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 2014;8(5):1280–9. https://doi.org/10.1016/j.celrep.2014.07.043.
Doyle GA, Crist RC, Karatas ET, Hammond MJ, Ewing AD, Ferraro TN, et al. Analysis of LINE-1 elements in DNA from postmortem brains of individuals with schizophrenia. Neuropsychopharmacology. 2017;42:2602–11. https://doi.org/10.1038/npp.2017.115. https://www.nature.com/articles/npp2017115#supplementary-information.
Guffanti G, Gaudi S, Klengel T, Fallon JH, Mangalam H, Madduri R, et al. LINE1 insertions as a genomic risk factor for schizophrenia: preliminary evidence from an affected family. Am J Med Genet B Neuropsychiatr Genet. 2016;171(4):534–45. https://doi.org/10.1002/ajmg.b.32437.
Shpyleva S, Melnyk S, Pavliv O, Pogribny I, Jill James S. Overexpression of LINE-1 retrotransposons in autism brain. Mol Neurobiol. 2018;55(2):1740–9. https://doi.org/10.1007/s12035-017-0421-x.
Liu S, Du T, Liu Z, Shen Y, Xiu J, Xu Q. Inverse changes in L1 retrotransposons between blood and brain in major depressive disorder. Sci Rep. 2016;6:37530. https://doi.org/10.1038/srep37530.
Evrony Gilad D, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann Hillel S, et al. Single-neuron sequencing analysis of L1 Retrotransposition and somatic mutation in the human brain. Cell. 2012;151(3):483–96. https://doi.org/10.1016/j.cell.2012.09.035.
Upton Kyle R, Gerhardt Daniel J, Jesuadian JS, Richardson Sandra R, Sánchez-Luque Francisco J, Bodea Gabriela O, et al. Ubiquitous L1 Mosaicism in Hippocampal Neurons. Cell. 2015;161(2):228–39. https://doi.org/10.1016/j.cell.2015.03.026.
Evrony GD, Lee E, Park PJ, Walsh CA. Resolving rates of mutation in the brain using single-neuron genomics. eLife. 2016;5:e12966. https://doi.org/10.7554/eLife.12966.
Lodato MA, Woodworth MB, Lee S, Evrony GD, Mehta BK, Karger A, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015;350(6256):94–8. https://doi.org/10.1126/science.aab1785.
• Lodato MA, Rodin RE, Bohrson CL, Coulter ME, Barton AR, Kwon M, et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science. 2018;359(6375):555. https://doi.org/10.1126/science.aao4426This study explores somatic single-nucleotide variations in individual neurons of the human brain in health and disease.
•• Chronister WD, Burbulis IE, Wierman MB, Wolpert MJ, Haakenson MF, Smith ACB, et al. Neurons with Complex Karyotypes Are Rare in Aged Human Neocortex. Cell Rep. 2019;26(4):825–35.e7. https://doi.org/10.1016/j.celrep.2018.12.107This study investigates the relationship between aging and somatic copy-number variations in neruons of the human brain.
Luo C, Keown CL, Kurihara L, Zhou J, He Y, Li J, et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science. 2017;357(6351):600–4. https://doi.org/10.1126/science.aan3351.
van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, et al. Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell. 2018;174(3):716–29.e27. https://doi.org/10.1016/j.cell.2018.05.061.
Li WV, Li JJ. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat Commun. 2018;9(1):997. https://doi.org/10.1038/s41467-018-03405-7.
Huang M, Wang J, Torre E, Dueck H, Shaffer S, Bonasio R, et al. SAVER: gene expression recovery for single-cell RNA sequencing. Nat Methods. 2018;15(7):539–42. https://doi.org/10.1038/s41592-018-0033-z.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–902.e21. https://doi.org/10.1016/j.cell.2019.05.031.
Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA, et al. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 2018;22(3):600–10. https://doi.org/10.1016/j.celrep.2017.12.072.
Gong T, Szustakowski JD. DeconRNASeq: a statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-Seq data. Bioinformatics (Oxford, England). 2013;29(8):1083–5. https://doi.org/10.1093/bioinformatics/btt090.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. https://doi.org/10.1038/nmeth.3337. https://www.nature.com/articles/nmeth.3337#supplementary-information.
Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar HA, et al. Genetic identification of brain cell types underlying schizophrenia. Nat Genet. 2018;50(6):825–33. https://doi.org/10.1038/s41588-018-0129-5.
Skene NG, Grant SGN. Identification of vulnerable cell types in major brain disorders using single cell transcriptomes and expression weighted cell type enrichment. Front Neurosci. 2016;10:16. https://doi.org/10.3389/fnins.2016.00016.
Calderon D, Bhaskar A, Knowles DA, Golan D, Raj T, Fu AQ, et al. Inferring relevant cell types for complex traits by using single-cell gene expression. Am J Hum Genet. 2017;101(5):686–99. https://doi.org/10.1016/j.ajhg.2017.09.009.
Codeluppi S, Borm LE, Zeisel A, La Manno G, van Lunteren JA, Svensson CI, et al. Spatial organization of the somatosensory cortex revealed by osmFISH. Nat Methods. 2018;15(11):932–5. https://doi.org/10.1038/s41592-018-0175-z.
Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018;362(6416):eaau5324. https://doi.org/10.1126/science.aau5324.
Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, et al. The human cell atlas. eLife. 2017;6:e27041. https://doi.org/10.7554/eLife.27041.
McConnell MJ, Moran JV, Abyzov A, Akbarian S, Bae T, Cortes-Ciriano I, et al. Intersection of diverse neuronal genomes and neuropsychiatric disease: the brain somatic mosaicism network. Science. 2017;356(6336):eaal1641. https://doi.org/10.1126/science.aal1641.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Malosree Maitra, Corina Nagy and Gustavo Turecki declare no conflicts of interest relevant to this manuscript.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Genetics and Neuroscience
Rights and permissions
About this article
Cite this article
Maitra, M., Nagy, C. & Turecki, G. Sequencing the Human Brain at Single-Cell Resolution. Curr Behav Neurosci Rep 6, 197–208 (2019). https://doi.org/10.1007/s40473-019-00192-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40473-019-00192-3