Abstract
Purpose of Review
We sought to (1) examine common sources of measurement error in research using data from electronic medical records (EMR), (2) discuss methods to assess the extent and type of measurement error, and (3) describe recent developments in methods to address this source of bias.
Recent Findings
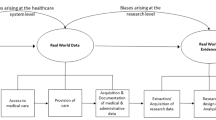
We identified eight sources of measurement error frequently encountered in EMR studies, the most prominent being that EMR data usually reflect only the health services and medications delivered within the specific health facility/system contributing to the EMR data. Methods for assessing measurement error in EMR data usually require gold standard or validation data, which may be possible using data linkage. Recent methodological developments to address the impact of measurement error in EMR analyses were particularly rich in the multiple imputation literature.
Summary
Presently, sources of measurement error impacting EMR studies are still being elucidated, as are methods for assessing and addressing them. Given the magnitude of measurement error that has been reported, investigators are urged to carefully evaluate and rigorously address this potential source of bias in studies based in EMR data.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major Importance
Wang LE, Shaw PA, Mathelier HM, Kimmel SE, French B. Evaluating risk-prediction models using data from electronic health records. Ann Appl Stat. 2016;10(1):286–304. https://doi.org/10.1214/15-aoas891.
•• Lin KJ, Glynn RJ, Singer DE, Murphy SN, Lii J, Schneeweiss S. Out-of-system care and recording of patient characteristics critical for comparative effectiveness research. Epidemiology. 2018;29(3):356–63. https://doi.org/10.1097/ede.0000000000000794. The authors use EMR data from two medical care networks linked with Medicare insurance claims to develop and assess data capture in EMR for 40 research-relevant variables. They report reporting surprisingly low capture proportions (16-27%), and propose a method to restrict EMR studies to patients with sufficiently informative data continuity.
Lin KJ, Singer DE, Glynn RJ, Murphy SN, Lii J, Schneeweiss S. Identifying patients with high data completeness to improve validity of comparative effectiveness research in electronic health records data. Clin Pharmacol Ther. 2018;103(5):899–905. https://doi.org/10.1002/cpt.861.
•• Weber GM, Adams WG, Bernstam EV, Bickel JP, Fox KP, Marsolo K, et al. Biases introduced by filtering electronic health records for patients with “complete data”. J Am Med Inform Assoc. 2017;24(6):1134–41. https://doi.org/10.1093/jamia/ocx071. Using EMR data from 7 (PCORNet) hospitals and health systems and (un-linked) Aetna insurance claims, the authors assess the impact of applying combinations of 16 different “complete-data” filters within EMR and claims populations. The authors demonstrate how missing data restrictions can be tailored to study-specific needs allowing for optimization of trade-offs between bias and generalizability.
Wells BJ, Chagin KM, Nowacki AS, Kattan MW. Strategies for handling missing data in electronic health record derived data. EGEMS (Wash DC). 2013;1(3):1035. https://doi.org/10.13063/2327-9214.1035.
Mooney SJ. Invited commentary: the tao of clinical cohort analysis-when the transitions that can be spoken of are not the true transitions. Am J Epidemiol. 2017;185(8):636–8. https://doi.org/10.1093/aje/kww236.
Fischer MA, Stedman MR, Lii J, Vogeli C, Shrank WH, Brookhart MA, et al. Primary medication non-adherence: analysis of 195,930 electronic prescriptions. J Gen Intern Med. 2010;25(4):284–90. https://doi.org/10.1007/s11606-010-1253-9.
Li X, Cole SR, Westreich D, Brookhart MA. Primary non-adherence and the new-user design. Pharmacoepidemiol Drug Saf. 2018;27(4):361–4. https://doi.org/10.1002/pds.4403.
Hampp C, Greene P, Pinheiro SP. Use of prescription drug samples in the USA: a descriptive study with considerations for pharmacoepidemiology. Drug Saf. 2016;39(3):261–70. https://doi.org/10.1007/s40264-015-0382-9.
Bijlsma MJ, Janssen F, Hak E. Estimating time-varying drug adherence using electronic records: extending the proportion of days covered (PDC) method. Pharmacoepidemiol Drug Saf. 2016;25(3):325–32. https://doi.org/10.1002/pds.3935.
•• Pye SR, Sheppard T, Joseph RM, Lunt M, Girard N, Haas JS, et al. Assumptions made when preparing drug exposure data for analysis have an impact on results: an unreported step in pharmacoepidemiology studies. Pharmacoepidemiol Drug Saf. 2018. https://doi.org/10.1002/pds.4440. Intended to clarify complex decision-making when defining drug treatment episodes in longitudinal data, the authors lay out a detailed algorithm/framework comprised of 10 decision nodes and 54 possible assumptions. They explore how variation in different decisions can impact effect estimates in an applied analysis conducted within UK CPRD data.
Pazzagli L, Linder M, Zhang M, Vago E, Stang P, Myers D, et al. Methods for time-varying exposure related problems in pharmacoepidemiology: an overview. Pharmacoepidemiol Drug Saf. 2018;27(2):148–60. https://doi.org/10.1002/pds.4372.
Devarakonda MV, Mehta N, Tsou CH, Liang JJ, Nowacki AS, Jelovsek JE. Automated problem list generation and physicians perspective from a pilot study. Int J Med Inform. 2017;105:121–9. https://doi.org/10.1016/j.ijmedinf.2017.05.015.
Zhang R, SVS P, Arsoniadis EG, Lee JT, Wang Y, Melton GB. Detecting clinically relevant new information in clinical notes across specialties and settings. BMC Med Inform Decis Mak. 2017;17(Suppl 2):68. https://doi.org/10.1186/s12911-017-0464-y.
Hubbard RA, Johnson E, Chubak J, Wernli KJ, Kamineni A, Bogart A, et al. Accounting for misclassification in electronic health records-derived exposures using generalized linear finite mixture models. Health Serv Outcome Res Methodol. 2017;17(2):101–12. https://doi.org/10.1007/s10742-016-0149-5.
Kreimeyer K, Foster M, Pandey A, Arya N, Halford G, Jones SF, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: a systematic review. J Biomed Inform. 2017;73:14–29. https://doi.org/10.1016/j.jbi.2017.07.012.
McTaggart S, Nangle C, Caldwell J, Alvarez-Madrazo S, Colhoun H, Bennie M. Use of text-mining methods to improve efficiency in the calculation of drug exposure to support pharmacoepidemiology studies. Int J Epidemiol. 2018;47(2):617–24. https://doi.org/10.1093/ije/dyx264.
Munkhdalai T, Liu F, Yu H. Clinical relation extraction toward drug safety surveillance using electronic health record narratives: classical learning versus deep learning. JMIR Public Health Surveill. 2018;4(2):e29. https://doi.org/10.2196/publichealth.9361.
Hamon T, Grabar N. Linguistic approach for identification of medication names and related information in clinical narratives. J Am Med Inform Assoc. 2010;17(5):549–54. https://doi.org/10.1136/jamia.2010.004036.
• Debray TP, Vergouwe Y, Koffijberg H, Nieboer D, Steyerberg EW, Moons KG. A new framework to enhance the interpretation of external validation studies of clinical prediction models. J Clin Epidemiol. 2015;68(3):279–89. https://doi.org/10.1016/j.jclinepi.2014.06.018. The authors propose a three-step framework for validating prediction models / algorithms which characterizes model performance in context of differences between the validation and development populations. Transportability is indicated by strong performance, maintained across heterogeneous validation and development populations. They walk through the framework in an applied example.
Lanes S, Brown JS, Haynes K, Pollack MF, Walker AM. Identifying health outcomes in healthcare databases. Pharmacoepidemiol Drug Saf. 2015;24(10):1009–16. https://doi.org/10.1002/pds.3856.
Lesko CR, Jacobson LP, Althoff KN, Abraham AG, Gange SJ, Moore RD, et al. Collaborative, pooled and harmonized study designs for epidemiologic research: challenges and opportunities. Int J Epidemiol. 2018;47(2):654–68. https://doi.org/10.1093/ije/dyx283.
Lin KJ, Garcia Rodriguez LA, Hernandez-Diaz S. Systematic review of peptic ulcer disease incidence rates: do studies without validation provide reliable estimates? Pharmacoepidemiol Drug Saf. 2011;20(7):718–28. https://doi.org/10.1002/pds.2153.
Koller KR, Wilson AS, Asay ED, Metzger JS, Neal DE. Agreement between self-report and medical record prevalence of 16 chronic conditions in the Alaska EARTH study. J Prim Care Community Health. 2014;5(3):160–5. https://doi.org/10.1177/2150131913517902.
Nakamura Y, Sugawara T, Kawanohara H, Ohkusa Y, Kamei M, Oishi K. Evaluation of estimated number of influenza patients from national sentinel surveillance using the national database of electronic medical claims. Jpn J Infect Dis. 2015;68(1):27–9. https://doi.org/10.7883/yoken.JJID.2014.092.
Stewart AL, Lynch KJ. Identifying discrepancies in electronic medical records through pharmacist medication reconciliation. J Am Pharm Assoc (2003). 2012;52(1):59–66. https://doi.org/10.1331/JAPhA.2012.10123.
Wright A, AB MC, Hickman TT, Hilaire DS, Borbolla D, Bowes WA 3rd, et al. Problem list completeness in electronic health records: A multi-site study and assessment of success factors. Int J Med Inform. 2015;84(10):784–90. https://doi.org/10.1016/j.ijmedinf.2015.06.011.
Rothman KJ, Greenland S, Lash TL. Clinical epidemiolgy. In: Seigafuse S, Bierig L, editors. Modern epidemiology. 3rd ed. Philadelphia: Lippincott Williams & Wilkins; 2008. p. 643.
Gini R, Schuemie MJ, Mazzaglia G, Lapi F, Francesconi P, Pasqua A, et al. Automatic identification of type 2 diabetes, hypertension, ischaemic heart disease, heart failure and their levels of severity from Italian General Practitioners’ electronic medical records: a validation study. BMJ Open. 2016;6(12):e012413. https://doi.org/10.1136/bmjopen-2016-012413.
Funk MJ, Landi SN. Misclassification in administrative claims data: quantifying the impact on treatment effect estimates. Curr Epidemiol Rep. 2014;1(4):175–85. https://doi.org/10.1007/s40471-014-0027-z.
Rowan CG, Flory J, Gerhard T, Cuddeback JK, Stempniewicz N, Lewis JD, et al. Agreement and validity of electronic health record prescribing data relative to pharmacy claims data: a validation study from a US electronic health record database. Pharmacoepidemiol Drug Saf. 2017;26(8):963–72. https://doi.org/10.1002/pds.4234.
Flory JH, Roy J, Gagne JJ, Haynes K, Herrinton L, Lu C, et al. Missing laboratory results data in electronic health databases: implications for monitoring diabetes risk. J Comp Eff Res. 2017;6(1):25–32. https://doi.org/10.2217/cer-2016-0033.
Patorno E, Gopalakrishnan C, Franklin JM, Brodovicz KG, Masso-Gonzalez E, Bartels DB, et al. Claims-based studies of oral glucose-lowering medications can achieve balance in critical clinical parameters only observed in electronic health records. Diabetes Obes Metab. 2017. https://doi.org/10.1111/dom.13184.
Heintzman J, Bailey SR, Hoopes MJ, Le T, Gold R, O’Malley JP, et al. Agreement of Medicaid claims and electronic health records for assessing preventive care quality among adults. J Am Med Inform Assoc. 2014;21(4):720–4. https://doi.org/10.1136/amiajnl-2013-002333.
Devoe JE, Gold R, McIntire P, Puro J, Chauvie S, Gallia CA. Electronic health records vs medicaid claims: completeness of diabetes preventive care data in community health centers. Ann Fam Med. 2011;9(4):351–8. https://doi.org/10.1370/afm.1279.
Yang S, Hutcheon JA. Identifying outliers and implausible values in growth trajectory data. Ann Epidemiol. 2016;26(1):77–80.e1–2. https://doi.org/10.1016/j.annepidem.2015.10.002.
Shi J, Korsiak J, Roth DE. New approach for the identification of implausible values and outliers in longitudinal childhood anthropometric data. Ann Epidemiol. 2018;28(3):204–11.e3. https://doi.org/10.1016/j.annepidem.2018.01.007.
Corbin M, Haslett S, Pearce N, Maule M, Greenland S. A comparison of sensitivity-specificity imputation, direct imputation and fully Bayesian analysis to adjust for exposure misclassification when validation data are unavailable. Int J Epidemiol. 2017;46(3):1063–72. https://doi.org/10.1093/ije/dyx027.
Sturmer T, Schneeweiss S, Rothman KJ, Avorn J, Glynn RJ. Performance of propensity score calibration--a simulation study. Am J Epidemiol. 2007;165(10):1110–8. https://doi.org/10.1093/aje/kwm074.
Sturmer T, Schneeweiss S, Rothman KJ, Avorn J, Glynn RJ. Propensity score calibration and its alternatives. Am J Epidemiol. 2007;165(10):1122–3.
Arah OA. Bias analysis for uncontrolled confounding in the health sciences. Annu Rev Public Health. 2017;38:23–38. https://doi.org/10.1146/annurev-publhealth-032315-021644.
Vanderweele TJ, Arah OA. Bias formulas for sensitivity analysis of unmeasured confounding for general outcomes, treatments, and confounders. Epidemiology. 2011;22(1):42–52. https://doi.org/10.1097/EDE.0b013e3181f74493.
• Rudolph KE, Stuart EA. Using sensitivity analyses for unobserved confounding to address covariate measurement error in propensity score methods. Am J Epidemiol. 2018;187(3):604–13. https://doi.org/10.1093/aje/kwx248. The authors propose adaptations of two prominent methods for assessing the impact of unobserved confounders (propensity score calibration, VanderWeele and Arah’s bias formulas) to instead assess the impact of measurement error. They illustrate the methods in an applied example.
Safran C, Bloomrosen M, Hammond WE, Labkoff S, Markel-Fox S, Tang PC, et al. Toward a national framework for the secondary use of health data: an American Medical Informatics Association White Paper. J Am Med Inform Assoc. 2007;14(1):1–9. https://doi.org/10.1197/jamia.M2273.
Velentgas P, Dreyer NA, Nourjah P, Smith SR, Torchia MM, editors. Developing a protocol for observational comparative effectiveness research: a user’s guide. AHRQ Publication No. 12(13)-EHC099. Rockville, MD: Agency for Healthcare Research and Quality; 2013. http://www.effectivehealthcare.ahrq.gov/Methods-OCER.cfm.
Rothman KJ, Greenland S, Lash TL. Case-control studies. In: Modern epidemiology. 3rd ed. Philadelphia: Lippincott Williams & Wilkins; 2008. p. 121–127.
Brunelli SM, Gagne JJ, Huybrechts KF, Wang SV, Patrick AR, Rothman KJ, et al. Estimation using all available covariate information versus a fixed look-back window for dichotomous covariates. Pharmacoepidemiol Drug Saf. 2013;22(5):542–50. https://doi.org/10.1002/pds.3434.
Conover MM, Jonsson Funk M. Uniform vs. all-available look-backs to identify exclusion criteria in observational cohort studies. Pharmacoepidem Dr S. 2015; 24(Supp 1):S689. https://doi.org/10.1002/pds [Abstract]
Nakasian SS, Rassen JA, Franklin JM. Effects of expanding the look-back period to all available data in the assessment of covariates. Pharmacoepidemiol Drug Saf. 2017;26(8):890–9. https://doi.org/10.1002/pds.4210.
Conover MM, Sturmer T, Poole C, Glynn RJ, Simpson RJ Jr, Pate V, et al. Classifying medical histories in US Medicare beneficiaries using fixed vs all-available look-back approaches. Pharmacoepidemiol Drug Saf. 2018. https://doi.org/10.1002/pds.4435.
Lewin A, Brondeel R, Benmarhnia T, Thomas F, Chaix B. Attrition bias related to missing outcome data: a longitudinal simulation study. Epidemiology. 2018;29(1):87–95. https://doi.org/10.1097/ede.0000000000000755.
• Lesko CR, Edwards JK, Cole SR, Moore RD, Lau B. When to censor? Am J Epidemiol. 2018;187(3):623–32. https://doi.org/10.1093/aje/kwx281. Informative loss to follow-up is an extremely common form of measurement error affecting time-to-event EMR studies. The authors provide needed guidance on how to appropriately right-censor follow-up time for outcomes that can be identified only during observed encounters vs. outside of observed encounters.
Little RJ, Rubin DB. Statistical analysis with missing data. 2nd ed. Hoboken, New Jersey: Wiley & Sons; 2002. https://doi.org/10.1002/9781119013563.
Wooldridge JM. Inverse probability weighted estimation for general missing data problems. J Econ. 2007;141(2):1281–301. https://doi.org/10.1016/j.jeconom.2007.02.002.
Doidge JC. Responsiveness-informed multiple imputation and inverse probability-weighting in cohort studies with missing data that are non-monotone or not missing at random. Stat Methods Med Res. 2018;27(2):352–63. https://doi.org/10.1177/0962280216628902.
Shin T, Davison ML, Long JD. Maximum likelihood versus multiple imputation for missing data in small longitudinal samples with nonnormality. Psychol Methods. 2017;22(3):426–49. https://doi.org/10.1037/met0000094.
Sun B, Perkins NJ, Cole SR, Harel O, Mitchell EM, Schisterman EF, et al. Inverse-probability-weighted estimation for monotone and nonmonotone missing data. Am J Epidemiol. 2018;187(3):585–91. https://doi.org/10.1093/aje/kwx350.
Rubin DB. Multiple imputation for nonresponse in surveys. Hoboken, New Jersey: Wiley & Sons; 2004. https://doi.org/10.1002/9780470316696.
Harel O, Mitchell EM, Perkins NJ, Cole SR, Tchetgen Tchetgen EJ, Sun B, et al. Multiple imputation for incomplete data in epidemiologic studies. Am J Epidemiol. 2018;187(3):576–84. https://doi.org/10.1093/aje/kwx349.
Schafer JL. Analysis of incomplete multivariate data. 1st ed. New York: Chapman and Hall/CRC; 1997. https://www.crcpress.com/Analysis-of-Incomplete-Multivariate-Data/Schafer/p/book/9781439821862.
Schafer JL. Multiple imputation: a primer. Stat Methods Med Res. 1999;8(1):3–15. https://doi.org/10.1177/096228029900800102.
Wei R, Wang J, Su M, Jia E, Chen S, Chen T, et al. Missing value imputation approach for mass spectrometry-based metabolomics data. Sci Rep. 2018;8(1):663. https://doi.org/10.1038/s41598-017-19120-0.
Dong Y, Peng CY. Principled missing data methods for researchers. Springerplus. 2013;2(1):222. https://doi.org/10.1186/2193-1801-2-222.
Rubin DB. Statistical matching using file concatenation with adjusted weights and multiple imputations. J Bus Econ Stat. 1986;4(1):87–94. https://doi.org/10.2307/1391390.
Rawlings AM, Sang Y, Sharrett AR, Coresh J, Griswold M, Kucharska-Newton AM, et al. Multiple imputation of cognitive performance as a repeatedly measured outcome. Eur J Epidemiol. 2017;32(1):55–66. https://doi.org/10.1007/s10654-016-0197-8.
Kunkel D, Kaizar EE. A comparison of existing methods for multiple imputation in individual participant data meta-analysis. Stat Med. 2017;36(22):3507–32. https://doi.org/10.1002/sim.7388.
Kline D, Andridge R, Kaizar E. Comparing multiple imputation methods for systematically missing subject-level data. Res Synth Methods. 2017;8(2):136–48. https://doi.org/10.1002/jrsm.1192.
Hill J. Reducing bias in treatment effect estimation in observational studies suffering from missing data. New York: Institute for Social and Economic Research and Policy, Columbia University; 2004. https://doi.org/10.7916/D8B85G11.
Mitra R, Reiter JP. A comparison of two methods of estimating propensity scores after multiple imputation. Stat Methods Med Res. 2016;25(1):188–204. https://doi.org/10.1177/0962280212445945.
Leyrat C, Seaman SR, White IR, Douglas I, Smeeth L, Kim J, et al. Propensity score analysis with partially observed covariates: how should multiple imputation be used? Stat Methods Med Res. 2017:962280217713032. https://doi.org/10.1177/0962280217713032.
Zahid FM, Heumann C. Multiple imputation with sequential penalized regression. Stat Methods Med Res. 2018:962280218755574. https://doi.org/10.1177/0962280218755574.
Sterne JA, White IR, Carlin JB, Spratt M, Royston P, Kenward MG, et al. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ. 2009;338:b2393.
Lee KJ, Carlin JB. Multiple imputation in the presence of non-normal data. Stat Med. 2017;36(4):606–17. https://doi.org/10.1002/sim.7173.
Geraci M, McLain A. Multiple imputation for bounded variables. Psychometrika. 2018. https://doi.org/10.1007/s11336-018-9616-y.
Sullivan TR, Lee KJ, Ryan P, Salter AB. Multiple imputation for handling missing outcome data when estimating the relative risk. BMC Med Res Methodol. 2017;17(1):134. https://doi.org/10.1186/s12874-017-0414-5.
• Bak N, Hansen LK. Data driven estimation of imputation error-a strategy for imputation with a reject option. PLoS One. 2016;11(10):e0164464. https://doi.org/10.1371/journal.pone.0164464. The authors describe a novel imputation method that selectively imputes values when they fall below a maximum error threshold. The method assesses imputation error among those with complete data, then assigns the error value to a person with missing data, who is non-parametrically matched using machine-learning.
Moreno-Betancur M, Carlin JB, Brilleman SL, Tanamas SK, Peeters A, Wolfe R. Survival analysis with time-dependent covariates subject to missing data or measurement error: multiple imputation for joint modeling (MIJM). Biostatistics. 2017. https://doi.org/10.1093/biostatistics/kxx046.
Kontopantelis E, Parisi R, Springate DA, Reeves D. Longitudinal multiple imputation approaches for body mass index or other variables with very low individual-level variability: the mibmi command in Stata. BMC Res Notes. 2017;10(1):41. https://doi.org/10.1186/s13104-016-2365-z.
Gottfredson NC, Sterba SK, Jackson KM. Explicating the conditions under which multilevel multiple imputation mitigates bias resulting from random coefficient-dependent missing longitudinal data. Prev Sci. 2017;18(1):12–9. https://doi.org/10.1007/s11121-016-0735-3.
Thompson CA, Boothroyd DB, Hastings KG, Cullen MR, Palaniappan LP, Rehkopf DH. A multiple-imputation “forward bridging” approach to address changes in the classification of asian race/ethnicity on the us death certificate. Am J Epidemiol. 2018;187(2):347–57. https://doi.org/10.1093/aje/kwx215.
Little RJ. Missing-data adjustments in large surveys. J Bus Econ Stat. 1988;6(3):287–96. https://doi.org/10.1080/07350015.1988.10509663.
Gu C, Gutman R. Combining item response theory with multiple imputation to equate health assessment questionnaires. Biometrics. 2017;73(3):990–8. https://doi.org/10.1111/biom.12638.
Siddique J, Reiter JP, Brincks A, Gibbons RD, Crespi CM, Brown CH. Multiple imputation for harmonizing longitudinal non-commensurate measures in individual participant data meta-analysis. Stat Med. 2015;34(26):3399–414. https://doi.org/10.1002/sim.6562.
Schomaker M, Heumann C. Bootstrap inference when using multiple imputation. Stat Med. 2018. https://doi.org/10.1002/sim.7654.
van Walraven C. Improved correction of misclassification bias with bootstrap imputation. Med Care. 2017. https://doi.org/10.1097/mlr.0000000000000787.
Wang C, Chen HY. Augmented inverse probability weighted estimator for Cox missing covariate regression. Biometrics. 2001;57(2):414–9. https://doi.org/10.1111/j.0006-341X.2001.00414.x.
• Hsu CH, Yu M. Cox regression analysis with missing covariates via nonparametric multiple imputation. Stat Methods Med Res. 2018:962280218772592. https://doi.org/10.1177/0962280218772592. The authors develop a novel method for addressing missing data for multiple covariates in time-to-event analysis that combines two existing methods: augmented inverse probability of treatment weighting (AIPW) and predictive mean matching imputation. The method is doubly-robust to model misspecification and is non-parametric, so suitable for non-normally distributed data.
Zhou M, He Y, Yu M, Hsu CH. A nonparametric multiple imputation approach for missing categorical data. BMC Med Res Methodol. 2017;17(1):87. https://doi.org/10.1186/s12874-017-0360-2.
Gardarsdottir H, Souverein PC, Egberts TC, Heerdink ER. Construction of drug treatment episodes from drug-dispensing histories is influenced by the gap length. J Clin Epidemiol. 2010;63(4):422–7. https://doi.org/10.1016/j.jclinepi.2009.07.001.
Hallas J, Gaist D, Bjerrum L. The waiting time distribution as a graphical approach to epidemiologic measures of drug utilization. Epidemiology. 1997;8(6):666–70. http://www.jstor.org/stable/3702660.
Pottegard A, Hallas J. Assigning exposure duration to single prescriptions by use of the waiting time distribution. Pharmacoepidemiol Drug Saf. 2013;22(8):803–9. https://doi.org/10.1002/pds.3459.
•• Støvring H, Pottegård A, Hallas J. Refining estimates of prescription durations by using observed covariates in pharmacoepidemiological databases: an application of the reverse waiting time distribution. Pharmacoepidemiol Drug Saf. 2017;26(8):900–8. https://doi.org/10.1002/pds.4216. The authors develop and apply a novel method, adapted from the reverse-waiting time distribution method, to estimate prescription durations in longitudinal data, modeled as a function of patient characteristics. Their data-driven method is more scalable and may be more accurate than the existing practice of specifying decision rules.
Støvring H, Pottegård A, Hallas J. Estimating medication stopping fraction and real-time prevalence of drug use in pharmaco-epidemiologic databases. An application of the reverse waiting time distribution. Pharmacoepidemiol Drug Saf. 2017;26(8):909–16. https://doi.org/10.1002/pds.4217.
Hallas J, Pottegard A, Stovring H. Using probability of drug use as independent variable in a register-based pharmacoepidemiological cause-effect study-an application of the reverse waiting time distribution. Pharmacoepidemiol Drug Saf. 2017;26(12):1520–6. https://doi.org/10.1002/pds.4326.
Ertefaie A, Flory JH, Hennessy S, Small DS. Instrumental variable methods for continuous outcomes that accommodate nonignorable missing baseline values. Am J Epidemiol. 2017;185(12):1233–9. https://doi.org/10.1093/aje/kww137.
Ertefaie A, Small DS, Flory JH, Hennessy S. A tutorial on the use of instrumental variables in pharmacoepidemiology. Pharmacoepidemiol Drug Saf. 2017;26(4):357–67. https://doi.org/10.1002/pds.4158.
Gault N, Castaneda-Sanabria J, De Rycke Y, Guillo S, Foulon S, Tubach F. Self-controlled designs in pharmacoepidemiology involving electronic healthcare databases: a systematic review. BMC Med Res Methodol. 2017;17(1):25. https://doi.org/10.1186/s12874-016-0278-0.
Funding
Dr. Jonsson Funk, Dr. Conover, and Ms. Young report grant support from the NIH National Institute on Aging (R01 AG056479), NIH National Heart Lung and Blood Institute (R01 HL118255), and NIH National Center for Advancing Translational Sciences (UL1 TR001111).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Pharmacoepidemiology
Rights and permissions
About this article
Cite this article
Young, J.C., Conover, M.M. & Jonsson Funk, M. Measurement Error and Misclassification in Electronic Medical Records: Methods to Mitigate Bias. Curr Epidemiol Rep 5, 343–356 (2018). https://doi.org/10.1007/s40471-018-0164-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40471-018-0164-x