Abstract
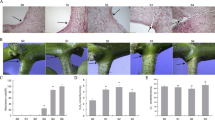
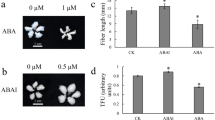
Leaves are the most important organs for photosynthesis. Many cotton leaf mutants (Gossypium spp.) have been reported and broadened the insights of leaf development. A natural occurring small, crinkled leaf (scl) mutant of upland cotton (Gossypium hirsutum L.) occurs in SY321 (wild type, WT), a high-yield upland cotton cultivar that is widely planted in China. The scl has a small and crinkled leaf phenotype. The shoot apical meristem (SAM) of scl did not form the essential typical layered structure, and the cell layers were arranged in a disordered manner. The free IAA concentration of shoot tips was significantly reduced in the scl according to ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) analysis. Gene expression profiles showed that 432 genes were differentially expressed between scl and WT. The top 20 enriched pathways were analyzed using the Kyoto encyclopedia of genes and genomes (KEGG) database and four pathways, including flavonoid biosynthesis, photosynthesis, RNA transport, and carbon fixation in photosynthetic organisms, were considered as significantly enriched pathways. Furthermore, a number of differentially expressed genes related to auxin activity, including flavonoids biosynthesis, auxin perception and signal transduction pathway, auxin transport, and ubiquitin-mediated proteolysis pathways, were identified, and the altered expressions of these genes might be responsible for the aberrant phenotype of the scl.







Similar content being viewed by others
References
Attia KA et al (2009) Antisense phenotypes reveal a functional expression of OsARF1, an auxin response factor, in transgenic rice. Curr Issues Mol Biol 11:i29–i34
Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7:986–995
Berleth T, Scarpella E, Prusinkiewicz P (2007) Towards the systems biology of auxin-transport-mediated patterning. Trends Plant Sci 12:151–159
Boggs JL, Tsegaye DTD, Coleman TL, Reddy KC, Fahsi A (2003) Relationship between hyperspectral reflectance, soil nitrate-nitrogen, cotton leaf chlorophyll, and cotton yield: a step toward precision agriculture. J Sustain Agric 22:5–16
Buer CS, Kordbacheh F, Truong TT, Hocart CH, Djordjevic MA (2013) Alteration of flavonoid accumulation patterns in transparent testa mutants disturbs auxin transport, gravity responses, and imparts long-term effects on root and shoot architecture. Planta 238:171–189
Calderon-Villalobos LI et al (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8:477–485
Chapman EJ, Estelle M (2009) Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 43:265–285
Cho M, Cho HT (2013) The function of ABCB transporters in auxin transport. Plant Signal Behav 8:e22990
Cui X, Guo Z, Song L, Wang Y, Cheng Y (2016) NCP1/AtMOB1A plays key roles in auxin-mediated arabidopsis development. PLoS Genet 12:e1005923
Dai JL, Dong HZ, Wei-Jiang LI (2009) Leaf senescence differences between cotton variety SGK 321 and SY 321 without chemical control of second generation bollworm. Shandong Agric Sci 2:19–22
Dewanto V, Wu X, Adom KK, Liu RH (2002) Thermal processing enhances the nutritional value of tomatoes by increasing total antioxidant activity. J Agric Food Chem 50:3010–3014
Doughty J, Aljabri M, Scott RJ (2014) Flavonoids and the regulation of seed size in Arabidopsis. Biochem Soc Trans 42:364–369
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38:W64–W70
Enders TA, Strader LC (2015) Auxin activity: past, present, and future. Am J Bot 102:180–196
Fambrini M et al (2006) Stem fasciated, a recessive mutation in sunflower (Helianthus annuus), alters plant morphology and auxin level. Ann Bot 98:715–730
Friml J (2003) Auxin transport—shaping the plant. Curr Opin Plant Biol 6:7–12
Fu J, Chu J, Sun X, Wang J, Yan C (2012) Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal Sci 28:1081–1087
Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y (2015) Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci USA 112:2275–2280
Gibson UE, Heid CA, Williams PM (1996) A novel method for real time quantitative RT-PCR. Genome Res 6:995–1001
Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414:271–276
Grunewald W et al (2012) Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc Natl Acad Sci USA 109:1554–1559
Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10:453–460
Havens KA, Guseman JM, Jang SS, Pierre-Jerome E, Bolten N, Klavins E, Nemhauser JL (2012) A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol 160:135–142
Heid CA, Stevens J, Livak KJ, Williams PM (1996) Real time quantitative PCR. Genome Res 6:986–994
Hutchinson JB (1946) The crinkled dwarf allelomorph series in the New World cottons. J Genet 47:178–207
Ichihashi Y, Tsukaya H (2015) Behavior of leaf meristems and their modification. Front Plant Sci 6:1060
Korasick DA, Enders TA, Strader LC (2013) Auxin biosynthesis and storage forms. J Exp Bot 64:2541–2555
Kuhn BM, Geisler M, Bigler L, Ringli C (2011) Flavonols accumulate asymmetrically and affect auxin transport in Arabidopsis. Plant Physiol 156:585–595
Kuhn BM et al (2016) 7-Rhamnosylated flavonols modulate homeostasis of the plant hormone auxin and affect plant development. J Biol Chem 291:5385–5395
Li F, Di J, Zhao L, Chen X (2014) Mapping of a new wrinkled leaf (wr3) gene in upland cotton. Yi Chuan 36:1256–1260
Li G, Zhu C, Gan L, Ng D, Xia K (2015) GA(3) enhances root responsiveness to exogenous IAA by modulating auxin transport and signalling in Arabidopsis. Plant Cell Rep 34:483–494
Liang W, Fang L, Xiang D, Hu Y, Feng H, Chang L, Zhang T (2015) Transcriptome analysis of short fiber mutant ligon lintless-1 (Li1) reveals critical genes and key pathways in cotton fiber elongation and leaf development. PLoS ONE 10:e0143503
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408
Mellor N et al (2016) Dynamic regulation of auxin oxidase and conjugating enzymes AtDAO1 and GH3 modulates auxin homeostasis. Proc Natl Acad Sci USA 113:11022–11027
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Ng JL, Hassan S, Truong TT, Hocart CH, Laffont C, Frugier F, Mathesius U (2015) Flavonoids and auxin transport inhibitors rescue symbiotic nodulation in the medicago truncatula cytokinin perception mutant cre1. Plant Cell 27:2210–2226
Noguero M et al (2015) DASH transcription factor impacts Medicago truncatula seed size by its action on embryo morphogenesis and auxin homeostasis. Plant J 81:453–466
Noh B, Murphy AS, Spalding EP (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13:2441–2454
Peer WA, Murphy AS (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12:556–563
Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS (2001) Flavonoid accumulation patterns of transparent testa mutants of arabidopsis. Plant Physiol 126:536–548
Petrasek J et al (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312:914–918
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Provenzano M, Mocellin S (2007) Complementary techniques: validation of gene expression data by quantitative real time PCR. Adv Exp Med Biol 593:66–73
Qu X, Zhao Z, Tian Z (2017) ERECTA regulates cell elongation by activating auxin biosynthesis in Arabidopsis thaliana. Front Plant Sci 8:1688
Saini S, Sharma I, Kaur N, Pati PK (2013) Auxin: a master regulator in plant root development. Plant Cell Rep 32:741–757
Salehin M, Bagchi R, Estelle M (2015) SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27:9–19
Shi CY et al (2011) Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genom 12:131
Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8:659–671
Singh RP, Singh VP, Singh M, Gupta GP, Lal CB (2001) Selection of narrow leaf upland cotton (Gossypium hirsutum) lines with better fibre quality and tolerance to whitefly (Bemisia tabaci). Indian J Agric Sci 71:131–133
Spiegelman Z, Omer S, Mansfeld BN, Wolf S (2017) Function of Cyclophilin1 as a long-distance signal molecule in the phloem of tomato plants. J Exp Bot 68:953–964
Steenackers W et al (2016) The allelochemical MDCA inhibits lignification and affects Auxin homeostasis. Plant Physiol 172:874–888
Strader LC, Zhao Y (2016) Auxin perception and downstream events. Curr Opin Plant Biol 33:8–14
Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446:640–645
Tang W et al (2014) The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol 202:509–520
Tivendale ND, Ross JJ, Cohen JD (2014) The shifting paradigms of auxin biosynthesis. Trends Plant Sci 19:44–51
Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15:533–543
Weijers D et al (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24:1874–1885
Xu T et al (2014) Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 343:1025–1028
Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E (2006) High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 16:1123–1127
Acknowledgements
This work was supported by the Grants from the National Key Research and Development Program of China (2016YFD0100203), National Science Foundation in China (31501351), National Project of Modern Agricultural Industry Technology System in China (CARS-15-05), Taishan Scholars Program of Shandong Province (No. ts201511070), and The Youth Scientific Research Foundation of Shandong Academy of Agricultural Science (2015YQN04).
Author information
Authors and Affiliations
Contributions
FW, JZ and JXZ designed the experiments, JXZ and YG carried out the lab experiments, CZ, GL and YC contributed to data analysis, FW, JZ and JXZ contributed to writing the paper.
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, J., Gao, Y., Zhang, C. et al. Auxin homeostasis and signaling alterations result in the aberrant phenotype in scl mutant of cotton (Gossypium hirsutum L.). Braz. J. Bot 41, 775–784 (2018). https://doi.org/10.1007/s40415-018-0493-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-018-0493-5