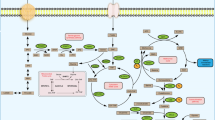
Abstract
Cellular metabolism reprogramming is an emerging hallmark of cancer, which provides tumor cells with not only necessary energy but also crucial materials to support growth. Exploiting the unique features of cancer metabolism is promising in cancer therapies. The growing interest in this field has led to numerous inhibitors being developed against key molecules in metabolic pathways, though most of them are still in preclinical development. Potential targeted cancer cell metabolic pathways under investigation include glycolysis, tricarboxylic acid (TCA) cycle, oxidative phosphorylation (OXPHOS), glutaminolysis, pentose phosphate pathway (PPP), lipid synthesis, amino acid and nucleotide metabolism. Sarcoma is a type of cancer that arises from transformed cells of mesenchymal origin, in contrast to carcinoma which originates from epithelial cells. Compared with carcinoma, progress towards harnessing the therapeutic potential of targeting sarcoma cell metabolism has been relatively slow. Recently however, with the discovery of cancer-specific mutations in metabolic enzymes such as isocitrate dehydrogenase (IDH) and succinate dehydrogenase (SDH) in certain sarcoma types, cancer cellular metabolism has been considered more as a source of new targets for treating sarcoma. In this article, we review metabolic enzymes currently tested for cancer therapies and describe the therapeutic potential of targeting IDH mutations and SDH deficiency in sarcomas.


Similar content being viewed by others
References
Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–33.
Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13(6):472–82.
Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18(1):54–61.
Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–7.
DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20.
Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23(5):537–48.
Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G. Metabolic targets for cancer therapy. Nat Rev Drug Discov. 2013;12(11):829–46.
Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol. 2017;14(1):11–31.
Gallamini A, Zwarthoed C, Borra A. Positron emission tomography (PET) in oncology. Cancers. 2014;6(4):1821–89.
Mankoff DA, Eary JF, Link JM, Muzi M, Rajendran JG, Spence AM, et al. Tumor-specific positron emission tomography imaging in patients: [18F] fluorodeoxyglucose and beyond. Clin Cancer Res. 2007;13(12):3460–9.
Dwarakanath BS, Singh D, Banerji AK, Sarin R, Venkataramana NK, Jalali R, et al. Clinical studies for improving radiotherapy with 2-deoxy-d-glucose: present status and future prospects. J Cancer Res Ther. 2009;5(Suppl 1):S21–6.
Papaldo P, Lopez M, Cortesi E, Cammilluzzi E, Antimi M, Terzoli E, et al. Addition of either lonidamine or granulocyte colony-stimulating factor does not improve survival in early breast cancer patients treated with high-dose epirubicin and cyclophosphamide. J Clin Oncol. 2003;21(18):3462–8.
Goldin N, Arzoine L, Heyfets A, Israelson A, Zaslavsky Z, Bravman T, et al. Methyl jasmonate binds to and detaches mitochondria-bound hexokinase. Oncogene. 2008;27(34):4636–43.
Jae HJ, Chung JW, Park HS, Lee MJ, Lee KC, Kim HC, et al. The antitumor effect and hepatotoxicity of a hexokinase II inhibitor 3-bromopyruvate: in vivo investigation of intraarterial administration in a rabbit VX2 hepatoma model. Korean J Radiol. 2009;10(6):596–603.
Clem BF, O’Neal J, Tapolsky G, Clem AL, Imbert-Fernandez Y, Kerr DA, et al. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol Cancer Ther. 2013;12(8):1461–70.
Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230-U74.
Dang CV. PKM2 tyrosine phosphorylation and glutamine metabolism signal a different view of the Warburg effect. Sci Signal. 2009;2(97):pe75. doi:10.1126/scisignal.297pe75.
Gupta V, Bamezai RNK. Human pyruvate kinase M2: A multifunctional protein. Protein Sci. 2010;19(11):2031–44.
Michels J, Obrist F, Castedo M, Vitale I, Kroemer G. PARP and other prospective targets for poisoning cancer cell metabolism. Biochem Pharmacol. 2014;92(1):164–71.
Khan JA, Forouhar F, Tao X, Tong L. Nicotinamide adenine dinucleotide metabolism as an attractive target for drug discovery. Expert Opin Ther Tar. 2007;11(5):695–705.
Holen K, Saltz LB, Hollywood E, Burk K, Hanauske AR. The pharmacokinetics, toxicities, and biologic effects of FK866, a nicotinamide adenine dinucleotide biosynthesis inhibitor. Invest New Drug. 2008;26(1):45–51.
Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. P Natl Acad Sci USA. 2010;107(5):2037–42.
Mutz CN, Schwentner R, Aryee DNT, Bouchard EDJ, Mejia EM, Hatch GM, et al. EWS-FLI1 confers exquisite sensitivity to NAMPT inhibition in Ewing sarcoma cells. Oncotarget. 2017;8(15):24679–93.
Hirschhaeuser F, Sattler UGA, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. 2011;71(22):6921–5.
Walenta S, Mueller-Klieser WF. Lactate: mirror and motor of tumor malignancy. Semin Radiat Oncol. 2004;14(3):267–74.
Finbow ME, Harrison MA. The vacuolar H+-ATPase: a universal proton pump of eukaryotes. Biochem J. 1997;324:697–712.
Sennoune SR, Martinez-Zaguilan R. Vacuolar H+-ATPase signaling pathway in cancer. Curr Protein Pept Sc. 2012;13(2):152–63.
Michel V, Licon-Munoz Y, Trujillo K, Bisoffi M, Parra KJ. Inhibitors of vacuolar ATPase proton pumps inhibit human prostate cancer cell invasion and prostate-specific antigen expression and secretion. Int J Cancer. 2013;132(2):E1–10.
Avnet S, Di Pompo G, Lemma S, Salerno M, Perut F, Bonuccelli G, et al. V-ATPase is a candidate therapeutic target for Ewing sarcoma. Bba Mol Basis Dis. 2013;1832(8):1105–16.
Perut F, Avnet S, Fotia C, Baglio SR, Salerno M, Hosogi S, et al. V-ATPase as an effective therapeutic target for sarcomas. Exp Cell Res. 2014;320(1):21–32.
Michelakis ED, Sutendra G, Dromparis P, Webster L, Haromy A, Niven E, et al. Metabolic Modulation of Glioblastoma with Dichloroacetate. Sci Transl Med. 2010;2(31). doi:10.1126/scitranslmed.3000677.
Michelakis ED, Webster L, Mackey JR. Dichloroacetate (DCA) as a potential metabolic-targeting therapy for cancer. Br J Cancer. 2008;99(7):989–94.
Papandreou I, Goliasova T, Denko NC. Anticancer drugs that target metabolism: is dichloroacetate the new paradigm? Int J Cancer. 2011;128(5):1001–8.
Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Power surge: Supporting cells “fuel” cancer cell mitochondria. Cell Metab. 2012;15(1):4–5.
Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov. 2010;9(6):447–64.
Barletta JA, Hornick JL. Succinate dehydrogenase-deficient tumors: diagnostic advances and clinical implications. Adv Anat Pathol. 2012;19(4):193–203.
Alam NA, Olpin S, Leigh IM. Fumarate hydratase mutations and predisposition to cutaneous leiomyomas, uterine leiomyomas and renal cancer. Br J Dermatol. 2005;153(1):11–7.
Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224(3):334–43.
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–44.
Abbas S, Lugthart S, Kavelaars FG, Schelen A, Koenders JE, Zeilemaker A, et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood. 2010;116(12):2122–6.
Hewitson KS, Lienard BM, McDonough MA, Clifton IJ, Butler D, Soares AS, et al. Structural and mechanistic studies on the inhibition of the hypoxia-inducible transcription factor hydroxylases by tricarboxylic acid cycle intermediates. J Biol Chem. 2007;282(5):3293–301.
Ponnaluri VK, Maciejewski JP, Mukherji M. A mechanistic overview of TET-mediated 5-methylcytosine oxidation. Biochem Biophys Res Commun. 2013;436(2):115–20.
Gottlieb E, Tomlinson IPM. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5(11):857–66.
Toro JR, Nickerson ML, Wei MH, Warren MB, Glenn GM, Turner ML, et al. Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America. Am J Hum Genet. 2003;73(1):95–106.
Ylisaukko-oja SK, Kiuru M, Lehtonen HJ, Lehtonen R, Pukkala E, Arola J, et al. Analysis of fumarate hydratase mutations in a population-based series of early onset uterine leiomyosarcoma patients. Int J Cancer. 2006;119(2):283–7.
Bo S, Ciccone G, Rosato R, Villois P, Appendino G, Ghigo E, et al. Cancer mortality reduction and metformin: a retrospective cohort study in type 2 diabetic patients. Diabetes Obes Metab. 2012;14(1):23–9.
Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330(7503):1304–5.
Soga T. Cancer metabolism: key players in metabolic reprogramming. Cancer Sci. 2013;104(3):275–81.
Bauer DE, Hatzivassiliou G, Zhao FP, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24(41):6314–22.
Hatzivassiliou G, Zhao FP, Bauer DE, Andreadis C, Shaw AN, Dhanak D, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8(4):311–21.
Anastasiou D, Cantley LC. Breathless cancer cells get fat on glutamine. Cell Res. 2012;22(3):443–6.
Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481(7381):380–4.
Filipp FV, Scott DA, Ronai ZA, Osterman AL, Smith JW. Reverse TCA cycle flux through isocitrate dehydrogenases 1 and 2 is required for lipogenesis in hypoxic melanoma cells. Pigm Cell Melanoma R. 2012;25(3):375–83.
DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29(3):313–24.
Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70(22):8981–7.
Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15(1):110–21.
Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014;13(4):890–901.
Pieter A, van den Heuvel J, Jing JP, Wooster RF, Bachman KE. Analysis of glutamine dependency in non-small cell lung cancer GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol Ther. 2012;13(12):1185–94.
Gameiro PA, Yang JJ, Metelo AM, Perez-Carro R, Baker R, Wang ZW, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 2013;17(3):372–85.
Riganti C, Gazzano E, Polimeni M, Aldieri E, Ghigo D. The pentose phosphate pathway: an antioxidant defense and a crossroad in tumor cell fate. Free Radic Bio Med. 2012;53(3):421–36.
Phan LM, Yeung SC, Lee MH. Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol Med. 2014;11(1):1–19.
Foldi M, Stickeler E, Bau L, Kretz O, Watermann D, Gitsch G, et al. Transketolase protein TKTL1 overexpression: a potential biomarker and therapeutic target in breast cancer. Oncol Rep. 2007;17(4):841–5.
Vizan P, Alcarraz-Vizan G, Diaz-Moralli S, Solovjeva ON, Frederiks WM, Cascante M. Modulation of pentose phosphate pathway during cell cycle progression in human colon adenocarcinoma cell line HT29. Int J Cancer. 2009;124(12):2789–96.
Xu XJ, Zur Hausen A, Coy JF, Lochelt M. Transketolase-like protein 1 (TKTL1) is required for rapid cell growth and full viability of human tumor cells. Int J Cancer. 2009;124(6):1330–7.
Yuan W, Wu S, Guo J, Chen Z, Ge J, Yang P, et al. Silencing of TKTL1 by siRNA inhibits proliferation of human gastric cancer cells in vitro and in vivo. Cancer Biol Ther. 2010;9(9):710–6.
Chajes V, Cambot M, Moreau K, Lenoir GM, Joulin V. Acetyl-CoA carboxylase alpha is essential to breast cancer cell survival. Cancer Res. 2006;66(10):5287–94.
Beckers A, Organe S, Tinunermans L, Scheys K, Peeters A, Brusselmans K, et al. Chemical inhibition of Acetyl-CoA carboxylase induces growth arrest and cytotoxicity selectively in cancer cells. Cancer Res. 2007;67(17):8180–7.
Flavin R, Peluso S, Nguyen PL, Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010;6(4):551–62.
Giro A, Blancafort A, Oliveras G, Perez-Bueno F, Dorca J, Vinas G, et al. Fatty acid synthase inhibition as a potential therapeutic target in triple-negative breast cancer. Eur J Cancer. 2013;49:S180-S.
Seguin F, Carvalho MA, Bastos DC, Agostini M, Zecchin KG, Alvarez-Flores MP, et al. The fatty acid synthase inhibitor orlistat reduces experimental metastases and angiogenesis in B16-F10 melanomas. Br J Cancer. 2012;107(6):977–87.
Uddin S, Hussain AR, Ahmed M, Abubaker J, Al-Sanea N, AbdulJabbar A, et al. High prevalence of fatty acid synthase expression in colorectal cancers in Middle Eastern patients and its potential role as a therapeutic target. Am J Gastroenterol. 2009;104(7):1790–801.
Kubatka P, Kruzliak P, Rotrekl V, Jelinkova S, Mladosievicova B. Statins in oncological research: from experimental studies to clinical practice. Crit Rev Oncol Hemat. 2014;92(3):296–311.
Nielsen SF, Nordestgaard BG, Bojesen SE. Statin use and reduced cancer-related mortality. New Engl J Med. 2012;367(19):1792–802.
Clem BF, Clem AL, Yalcin A, Goswami U, Arumugam S, Telang S, et al. A novel small molecule antagonist of choline kinase-alpha that simultaneously suppresses MAPK and PI3K/AKT signaling. Oncogene. 2011;30(30):3370–80.
Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer. 2011;11(12):835–48.
Chabner BA, Roberts TG. Timeline—chemotherapy and the war on cancer. Nat Rev Cancer. 2005;5(1):65–72.
Visentin M, Zhao RB, Goldman ID. The antifolates. Hematol Oncol Clin N. 2012;26(3):629–48.
Sabini E, Ort S, Monnerjahn C, Konrad M, Lavie A. Structure of human dCK suggests strategies to improve anticancer and antiviral therapy. Nat Struct Biol. 2003;10(7):513–9.
Braas D, Ahler E, Tam B, Nathanson D, Riedinger M, Benz MR, et al. Metabolomics strategy reveals subpopulation of liposarcomas sensitive to gemcitabine treatment. Cancer Discov. 2012;2(12):1109–17.
Lukey MJ, Katt WP, Cerione RA. Targeting amino acid metabolism for cancer therapy. Drug Discov Today. 2016. doi:10.1016/j.drudis.2016.12.003.
Qiu F, Huang J, Sui M. Targeting arginine metabolism pathway to treat arginine-dependent cancers. Cancer Lett. 2015;364(1):1–7.
Kremer JC, Prudner BC, Lange SE, Bean GR, Schultze MB, Brashears CB, et al. Arginine deprivation inhibits the Warburg effect and upregulates glutamine anaplerosis and serine biosynthesis in ASS1-deficient cancers. Cell Rep. 2017;18(4):991–1004.
Katz JB, Muller AJ, Prendergast GC. Indoleamine 2,3-dioxygenase in T-cell tolerance and tumoral immune escape. Immunol Rev. 2008;222:206–21.
Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348(6230):74–80.
Vacchelli E, Aranda F, Eggermont A, Sautes-Fridman C, Tartour E, Kennedy EP, et al. Trial watch: IDO inhibitors in cancer therapy. Oncoimmunology. 2014;3(10):e957994.
Okamoto A, Nikaido T, Ochiai K, Takakura S, Saito M, Aoki Y, et al. Indoleamine 2,3-dioxygenase serves as a marker of poor prognosis in gene expression profiles of serous ovarian cancer cells. Clin Cancer Res. 2005;11(16):6030–9.
Urakawa H, Nishida Y, Nakashima H, Shimoyama Y, Nakamura S, Ishiguro N. Prognostic value of indoleamine 2,3-dioxygenase expression in high grade osteosarcoma. Clin Exp Metastasis. 2009;26(8):1005–12.
Balachandran VP, Cavnar MJ, Zeng S, Bamboat ZM, Ocuin LM, Obaid H, et al. Imatinib potentiates antitumor T cell responses in gastrointestinal stromal tumor through the inhibition of Ido. Nat Med. 2011;17(9):1094–100.
Zhai L, Spranger S, Binder DC, Gritsina G, Lauing KL, Giles FJ, et al. Molecular pathways: targeting IDO1 and other tryptophan dioxygenases for cancer immunotherapy. Clin Cancer Res. 2015;21(24):5427–33.
Xu X, Zhao J, Xu Z, Peng B, Huang Q, Arnold E, et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279(32):33946–57.
Waitkus MS, Diplas BH, Yan H. Isocitrate dehydrogenase mutations in gliomas. Neuro Oncol. 2016;18(1):16–26.
Haselbeck RJ, McAlister-Henn L. Function and expression of yeast mitochondrial NAD- and NADP-specific isocitrate dehydrogenases. J Biol Chem. 1993;268(16):12116–22.
Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer I. 2010;102(13):932–41.
Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–53.
Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3(7):730–41.
Yen KE, Schenkein DP. Cancer-associated isocitrate dehydrogenase mutations. Oncol. 2012;17(1):5–8.
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–73.
Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–34.
Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207(2):339–44.
Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–12.
Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–66.
Zhang Y, Paz Mejia A, Temple HT, Trent J, Rosenberg AE. Squamous cell carcinoma arising in dedifferentiated chondrosarcoma proved by isocitrate dehydrogenase mutation analysis. Hum Pathol. 2014;45(7):1541–5.
Tinoco G, Wilky BA, Paz-Mejia A, Rosenberg A, Trent JC. The biology and management of cartilaginous tumors: a role for targeting isocitrate dehydrogenase. Am Soc Clin Oncol Educ Book Am Soc Clin Oncol Meet. 2015. doi:10.14694/EdBook_AM.2015.35.e648.
Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncol. 2012;17(1):72–9.
Wang P, Dong Q, Zhang C, Kuan PF, Liu Y, Jeck WR, et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32(25):3091–100.
Lopez GY, Reitman ZJ, Solomon D, Waldman T, Bigner DD, McLendon RE, et al. IDH1(R132) mutation identified in one human melanoma metastasis, but not correlated with metastases to the brain. Biochem Biophys Res Commun. 2010;398(3):585–7.
Shibata T, Kokubu A, Miyamoto M, Sasajima Y, Yamazaki N. Mutant IDH1 confers an in vivo growth in a melanoma cell line with BRAF mutation. Am J Pathol. 2011;178(3):1395–402.
Tang JY, Chang CC, Lin PC, Chang JG. Isocitrate dehydrogenase mutation hot spots in acute lymphoblastic leukemia and oral cancer. Kaohsiung J Med Sci. 2012;28(3):138–44.
Sequist LV, Heist RS, Shaw AT, Fidias P, Rosovsky R, Temel JS, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol. 2011;22(12):2616–24.
Murugan AK, Bojdani E, Xing M. Identification and functional characterization of isocitrate dehydrogenase 1 (IDH1) mutations in thyroid cancer. Biochem Biophys Res Commun. 2010;393(3):555–9.
Fathi AT, Sadrzadeh H, Comander AH, Higgins MJ, Bardia A, Perry A, et al. Isocitrate dehydrogenase 1 (IDH1) mutation in breast adenocarcinoma is associated with elevated levels of serum and urine 2-hydroxyglutarate. Oncol. 2014;19(6):602–7.
Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125(2):353–5.
Ghiam AF, Cairns RA, Thoms J, Dal Pra A, Ahmed O, Meng A, et al. IDH mutation status in prostate cancer. Oncogene. 2012;31(33):3826.
Gaal J, Burnichon N, Korpershoek E, Roncelin I, Bertherat J, Plouin PF, et al. Isocitrate dehydrogenase mutations are rare in pheochromocytomas and paragangliomas. J Clin Endocrinol Metab. 2010;95(3):1274–8.
Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais JP, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119(8):1901–3.
Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–9.
Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Bio. 2013;14(6):341–56.
Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–67.
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30.
Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–8.
Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–9.
Turcan S, Fabius AW, Borodovsky A, Pedraza A, Brennan C, Huse J, et al. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT Inhibitor Decitabine. Oncotarget. 2013;4(10):1729–36.
Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–6.
Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–30.
Kernytsky A, Wang F, Hansen E, Schalm S, Straley K, Gliser C, et al. IDH2 mutation-induced histone and DNA hypermethylation is progressively reversed by small-molecule inhibition. Blood. 2015;125(2):296–303.
Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529(7584):110–4.
Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483(7390):484–8.
Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324(5924):261–5.
Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339(6127):1621–5.
Losman JA, Kaelin WG. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27(8):836–52.
Cleven AHG, Suijker J, Agrogiannis G, Briaire-de Bruijn IH, Frizzell N, Hoekstra AS, et al. IDH1 or -2 mutations do not predict outcome and do not cause loss of 5-hydroxymethylcytosine or altered histone modifications in central chondrosarcomas. Clin Sarcoma Res. 2017;7:8. doi:10.1186/s13569-017-0074-6.
Suijker J, Baelde HJ, Roelofs H, Cleton-Jansen AM, Bovee JVMG. The oncometabolite D-2-hydroxyglutarate induced by mutant IDH1 or-2 blocks osteoblast differentiation in vitro and in vivo. Oncotarget. 2015;6(17):14832–42.
Hansen E, Quivoron C, Straley K, Lemieux RM, Popovici-Muller J, Sadrzadeh H, et al. AG-120, an Oral, Selective, First-in-Class, Potent Inhibitor of Mutant IDH1, Reduces Intracellular 2HG and Induces Cellular Differentiation in TF-1 R132H Cells and Primary Human IDH1 Mutant AML Patient Samples Treated Ex Vivo. Blood. 2014;124(21):3734.
Quivoron C, David M, Straley K, Travins J, Kim H, Chen Y, et al. AG-221, an Oral, Selective, First-in-Class, Potent IDH2-R140Q Mutant Inhibitor, Induces Differentiation in a Xenotransplant Model. Blood. 2014;124(21):3735.
Li L, Paz AC, Wilky BA, Johnson B, Galoian K, Rosenberg A, et al. Treatment with a small molecule mutant IDH1 inhibitor suppresses tumorigenic activity and decreases production of the oncometabolite 2-hydroxyglutarate in human chondrosarcoma cells. PLoS One. 2015;10(9):e0133813.
Chaturvedi A, Herbst L, Pusch S, Klett L, Goparaju R, Stichel D, et al. Pan-mutant-IDH1 inhibitor BAY1436032 is highly effective against human IDH1 mutant acute myeloid leukemia in vivo. Leukemia. 2017. doi:10.1038/leu.2017.46.
Evenepoel L, Papathomas TG, Krol N, Korpershoek E, de Krijger RR, Persu A, et al. Toward an improved definition of the genetic and tumor spectrum associated with SDH germ-line mutations. Genet Med. 2015;17(8):610–20.
Janeway KA, Kim SY, Lodish M, Nose V, Rustin P, Gaal J, et al. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. P Natl Acad Sci USA. 2011;108(1):314–8.
Ricketts C, Woodward ER, Killick P, Morris MR, Astuti D, Latif F, et al. Germline SDHB mutations and familial renal cell carcinoma. J Natl Cancer I. 2008;100(17):1260–2.
Miettinen M, Lasota J. Succinate dehydrogenase deficient gastrointestinal stromal tumors (GISTs)—a review. Int J Biochem Cell B. 2014;53:514–9.
Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer. 2011;11(12):865–78.
Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21(23):4342–9.
Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279(5350):577–80.
Wang YM, Gu ML, Ji F. Succinate dehydrogenase-deficient gastrointestinal stromal tumors. World J Gastroenterol. 2015;21(8):2303–14.
Weldon CB, Madenci AL, Boikos SA, Janeway KA, George S, von Mehren M, et al. Surgical management of wild-type gastrointestinal stromal tumors: a report from the national institutes of health pediatric and wildtype GIST clinic. J Clin Oncol. 2016;35(5):523–8.
Miettinen M, Wang ZF, Sarlomo-Rikala M, Osuch C, Rutkowski P, Lasota J. Succinate dehydrogenase-deficient GISTs: a clinicopathologic, immunohistochemical, and molecular genetic study of 66 gastric GISTs with predilection to young age. Am J Surg Pathol. 2011;35(11):1712–21.
Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7(1):77–85.
Antonescu CR, Viale A, Sarran L, Tschernyavsky SJ, Gonen M, Segal NH, et al. Gene expression in gastrointestinal stromal tumors is distinguished by KIT genotype and anatomic site. Clin Cancer Res. 2004;10(10):3282–90.
Chou A, Chen J, Clarkson A, Samra JS, Clifton-Bligh RJ, Hugh TJ, et al. Succinate dehydrogenase-deficient GISTs are characterized by IGF1R overexpression. Modern Pathol. 2012;25(9):1307–13.
Nannini M, Biasco G, Astolfi A, Pantaleo MA. An overview on molecular biology of KIT/PDGFRA wild type (WT) gastrointestinal stromal tumours (GIST). J Med Genet. 2013;50(10):653–61.
Pantaleo MA, Astolfi A, Di Battista M, Heinrich MC, Paterini P, Scotlandi K, et al. Insulin-like growth factor 1 receptor expression in wild-type GISTs: a potential novel therapeutic target. Int J Cancer. 2009;125(12):2991–4.
Corless CL, Beadling C, Justusson E, Heinrich MC. Evaluation of the presence of IGF1R overexpression in wild-type and kinase mutant GI stromal tumors. J Clin Oncol. 2009;27(15):10506.
Songdej N, von Mehren M. GIST treatment options after tyrosine kinase inhibitors. Curr Treat Options Oncol. 2014;15(3):493–506.
Pollak M. The insulin receptor/insulin-like growth factor receptor family as a therapeutic target in oncology. Clin Cancer Res. 2012;18(1):40–50.
Acknowledgements
We acknowledge partial support and assistance from the Sheila and David Fuente Graduate Program in Cancer Biology, Sylvester Comprehensive Cancer Center.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No funding was received for the preparation of this review.
Conflicts of interest
Luyuan Li, Josiane E. Eid, Ana C. Paz and Jonathan C. Trent declare no conflicts of interest.
Rights and permissions
About this article
Cite this article
Li, L., Eid, J.E., Paz, A.C. et al. Metabolic Enzymes in Sarcomagenesis: Progress Toward Biology and Therapy. BioDrugs 31, 379–392 (2017). https://doi.org/10.1007/s40259-017-0237-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40259-017-0237-2