Abstract
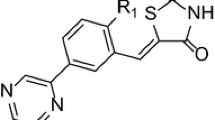
PIM2 kinase plays a crucial role in the cell cycle events including survival, proliferation, and differentiation in normal and neoplastic neuronal cells. Thus, it is regarded as an essential target for cancer pharmaceutical. Design of novel 5-(1H-indol-5-yl)-1,3,4-thiadiazol-2-amine derivatives with enhanced PIM2 inhibitory activity. A series of twenty-five PIM2 inhibitors reported in the literature containing 5-(1H-indol-5-yl)-1,3,4-thiadiazol-2-amines scaffold was studied by using two computational techniques, namely, three-dimensional quantitative structure activity relationship (3D-QSAR) and molecular docking. The comparative molecular field analysis (CoMFA) and comparative molecular similarity indexes analysis (CoMSIA) studies were developed using nineteen molecules having pIC50 ranging from 8.222 to 4.157. The best generated CoMFA and CoMSIA models exhibit conventional determination coefficients R2 of 0.91 and 0.90 as well as the Leave One Out cross-validation determination coefficients Q2 of 0.68 and 0.62, respectively. Moreover, the predictive ability of those models was evaluated by the external validation using a test set of six compounds with predicted determination coefficients R 2test of 0.96 and 0.96, respectively. Besides, y-randomization test was also performed to validate our 3D-QSAR models. The most and the least active compounds were docked into the active site of the protein (PDB ID: 4 × 7q) to confirm those obtained results from 3D-QSAR models and elucidate the binding mode between this kind of compounds and the PIM2 enzyme. These satisfactory results are not offered help only to understand the binding mode of 5-(1H-indol-5-yl)-1,3,4-thiadiazol series compounds into this kind of targets, but provide information to design new potent PIM2 inhibitors.










Similar content being viewed by others
References
AbdulHameed MDM, Hamza A, Liu J, Zhan C-G (2008) Combined 3D-QSAR modeling and molecular docking study on indolinone derivatives as inhibitors of 3-phosphoinositide-dependent protein kinase-1. J Chem Inf Model 48(9):1760–1772. https://doi.org/10.1021/ci800147v
Baroni M, Clementi S, Cruciani G, Costantino G, Oberrauch D, Riganelli E (1992) Predictive ability of regression models. Part II: selection of the best predictive PLS model. J Chemom 6:347–356
Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller J (2010) PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica [Internet] 95(6):1004–1015. Available from: http://www.haematologica.org/cgi/doi/10.3324/haematol.2009.017079
Bullock AN, Russo S, Amos A, Pagano N, Bregman H, Debreczeni JÉ et al (2009) Crystal structure of the PIM2 kinase in complex with an organoruthenium inhibitor. Gay N, editor. PLoS ONE 4(10):e7112. https://doi.org/10.1371/journal.pone.0007112
Clark M, Cramer RD, Van Opdenbosch N (1989) Validation of the general purpose tripos 5.2 force field. J Comput Chem [Internet] 10(8):982–1012. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12192139
Cruciani G, Baroni M, Clementi S, Costantino G, Riganelli D, Skagerberg B (1992) Predictive ability of regression models. Part I: standard deviation of prediction errors (SDEP). J Chemom 6(6):335–346. https://doi.org/10.1002/cem.1180060604
Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 2017, San Diego: Dassault Systèmes. [Internet]. [cited 2017 Feb 25]. Available from: http://accelrys.com/products/collaborative-science/biovia-discovery-studio/
Gadewal N, Varma A (2012) Targeting Pim-1 kinase for potential drug-development. Int J Comput Biol Drug Des 5(2):137–151
Golbraikh A, Tropsha A (2002) Beware of q2! J. Mol. Graph. Model [Internet] 20(4):269–76. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11858635
Gupta SP, Mathur AN, Nagappa AN, Kumar D, Kumaran S (2003) A quantitative structure-activity relationship study on a novel class of calcium-entry blockers: 1-[(4-(aminoalkoxy)phenyl)sulphonyl]indolizines. Eur J Med Chem [Internet]. 38(10):867–73. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14575933
Hong S, Kim J, Seo JH, Jung KH, Hong SS, Hong S (2012) Design, synthesis, and evaluation of 3,5-disubstituted 7-azaindoles as Trk inhibitors with anticancer and antiangiogenic activities. J Med Chem 55(11):5337–5349. https://doi.org/10.1021/jm3002982
Klebe G, Abraham U, Mietzner T (1994) Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J Med Chem 37(24):4130–4146. https://doi.org/10.1021/jm00050a010
Kubinyi H (2003) Comparative molecular field analysis (CoMFA). Handb Chemoinformatics. https://doi.org/10.1002/9783527618279.ch44d
Lipinski CA (2004) Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1(4):337–41. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1740674904000551
Nawijn MC, Alendar A, Berns A (2011a) For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer Nature Publishing Group 11(1):23–34. Available from: http://www.nature.com/nrc/journal/v11/n1/pdf/nrc2986.pdf%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/21150935
Nawijn MC, Alendar A, Berns A (2011b) For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer 11(1):23–34. https://doi.org/10.1038/nrc2986
Purcell WP, Singer JA (1967) A brief review and table of semiempirical parameters used in the Hueckel molecular orbital method. J Chem Eng Data 12(2):235–246. https://doi.org/10.1021/je60033a020
Qian K, Lian W, Cywin CL, Farmer BT, Hickey E, Homon C et al (2009) Hit to lead account of the discovery of a new class of inhibitors of pim kinases and crystallographic studies revealing an unusual kinase binding mode. J Med Chem 52(7):1814–1827
Rücker C, Rücker G, Meringer M (2007) Y-randomization and its variants in QSPR/QSAR. J Chem Inf Model 47(6):2345–2357
Santio NM, Vahakoski RL, Rainio E-M, Sandholm JA, Virtanen SS, Prudhomme M et al (2010) Pim-selective inhibitor DHPCC-9 reveals Pim kinases as potent stimulators of cancer cell migration and invasion. Mol Cancer 9(1):279. https://doi.org/10.1186/1476-4598-9-279
Ståhle L, Wold S (1988) Multivariate data analysis and experimental design in biomedical research. Prog Med Chem 25:291–338. Available from: http://www.ncbi.nlm.nih.gov/pubmed/3076969
SYBYL-X 2.0 [Internet]. St. Louis, MO, USA: Tripos Inc; Available from: http://www.tripos.com
Tropsha A, Gramatica P, Gombar VK (2003) The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. QSAR Comb Sci 22(1):69–77
Wold S (1991) Validation of QSAR’s. Quant Struct Relatsh 10(3):191–193. https://doi.org/10.1002/qsar.19910100302
Wu B, Wang HL, Cee VJ, Lanman BA, Nixey T, Pettus L et al (2015) Discovery of 5-(1H-indol-5-yl)-1,3,4-thiadiazol-2-amines as potent PIM inhibitors. Bioorganic Med Chem Lett 25(4):775–780. https://doi.org/10.1016/j.bmcl.2014.12.091
Wurz RP, Pettus LH, Jackson C, Wu B, Wang HL, Herberich B et al (2015) The discovery and optimization of aminooxadiazoles as potent Pim kinase inhibitors. Bioorganic Med Chem Lett 25(4):847–855. https://doi.org/10.1016/j.bmcl.2014.12.067
Zheng J, Xiao G, Guo J, Zheng Y, Gao H, Zhao S et al (2011) Exploring QSARs for 5-Lipoxygenase (5-LO) Inhibitory Activity of 2-Substituted 5-Hydroxyindole-3-Carboxylates by CoMFA and CoMSIA. Chem Biol Drug Des 78(2):314–321. https://doi.org/10.1111/j.1747-0285.2011.01146.x
Acknowledgment
We are grateful to the “Association Marocaine des Chimistes Théoriciens” (AMCT) and “Moroccan centre of scientific and technique research” (CNRST) for their pertinent help concerning the programs.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Aouidate, A., Ghaleb, A., Ghamali, M. et al. Computer aided drug design based on 3D-QSAR and molecular docking studies of 5-(1H-indol-5-yl)-1,3,4-thiadiazol-2-amine derivatives as PIM2 inhibitors: a proposal to chemists. In Silico Pharmacol. 6, 5 (2018). https://doi.org/10.1007/s40203-018-0043-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40203-018-0043-7