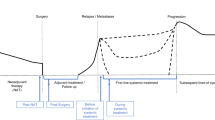
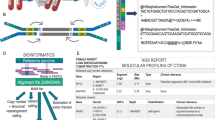
Abstract
The heterogeneity of tumor is considered as a major difficulty to victorious personalized cancer medicine. There is an extremeneed of consistent response evaluation for in vivo tumor heterogeneity anditscoupledconflict mechanisms. In this occasion researchers will be able to keep pace withpredictive, preventive, personalized, and Participatory (P4) medicine for cancer managements. In fact tumor heterogeneity is a central part of cancer evolution,soin order to progress in understanding of the dynamics within a tumor some diagnostic apparatus should be improved. Latest molecular techniques like Next generation Sequencing (NGS) and ultra-deep sequencing could disclose some clones within a liquid tumor biopsy which mainly responsible of treatment resistance. Circulating tumor DNA (ctDNA) as a main component of liquid biopsy is agifted biomarker for cancer mutation tracking as well as profiling. Personalized medicine facilitate learning regarding to genetic pools of tumor and their possible respond to treatment which could be much easier by using of ctDNA.With this information, cliniciansarelooking forward to find the best strategies for prevention, screening, and treatment in the way of precision medicine. Currently, numerous clinical efficacy of such informative improved treatment are in hand. Here we represent the review of plasma-derived ctDNA studies use in personalized cancer managements.

Similar content being viewed by others
Abbreviations
- ATC:
-
Anaplastic Thyroid Cancer
- AI:
-
Aromatase inhibitor
- BC:
-
Breast Cancer
- CAPP-Seq:
-
Cancer Personalized Profiling by deep Sequencing
- CRC:
-
Colorectal cancers
- ctDNA:
-
Circulating Tumor DNA
- CTCs:
-
Circulating Tumor Cells
- ddPCR:
-
Droplet Digital PCR
- EGFR-TKIs:
-
Epidermal growth factor receptor tyrosine kinase inhibitors
- EGFR:
-
Epidermal growth factor receptor
- ESR1:
-
Estrogen receptor alpha
- FDA:
-
U.S. Food and Drug Administration
- HER2/neu:
-
Human epidermal growth factor receptor 2
- KRAS:
-
Kirsten Rat Sarcoma Viral Oncogene Homolog
- mCRC:
-
Metastatic colorectal cancer
- MRD:
-
Minimal residual disease
- MTC:
-
Medularlly Thyroid Cancer
- NSCLC:
-
Non-Small Cell Lung Cancer Research
- NGS:
-
Next-generation sequencing
- PFS:
-
Progression free survival
- PIK3C:
-
Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha
- PI3K:
-
Phosphatidylinositol-4,5-bisphosphate 3-kinase
- PDGFRA:
-
Platelet-derived growth factor receptor alpha
References
Larijani B, Shirzad M, Mohagheghi M, Haghpanah V, Mosavi-Jarrahi A, Tavangar S, et al. Epidemiologic analysis of the Tehran cancer institute data system registry (TCIDSR). Asian Pac J Cancer Prev. 2004;5(1):36–9.
Haghpanah V, Soliemanpour B, Heshmat R, Mosavi-Jarrahi A, Tavangar S, Malekzadeh R, et al. Endocrine cancer in Iran: based on cancer registry system. Indian J Cancer. 2006;43(2):80–5.
Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, et al. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12(7):e1006162.
Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–37.
Stroun M, Anker P. Nucleic acids spontaneously released by living frog auricles. Biochem J. 1972;128(3):100P.
Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA: apoptosis and active DNA release. Clin Chim Acta. 2001;313(1):139–42.
Anker P, Stroun M, Maurice PA. Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system. Cancer Res. 1975;35(9):2375–82.
Rogers JC, Boldt D, Kornfeld S, Skinner SA, Valeri CR. Excretion of deoxyribonucleic acid by lymphocytes stimulated with phytohemagglutinin or antigen. Proc Natl Acad Sci. 1972;69(7):1685–9.
Diaz Jr LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–86.
Hamakawa T, Kukita Y, Kurokawa Y, Miyazaki Y, Takahashi T, Yamasaki M, et al. Monitoring gastric cancer progression with circulating tumour DNA. Br J Cancer. 2015;112(2):352.
Tavangar SM, Larijani B, Mahta A, Hosseini SMA, Mehrazine M, Bandarian F. Craniopharyngioma: a clinicopathological study of 141 cases. Endocr Pathol. 2004;15(4):339–44.
Pantel K, Alix-Panabières C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res. 2013;73(21):6384–8.
Costs AA. Outcomes comparison of tissue and blood based biopsies for the purpose of biomarker testing. Value Health. 2016;19(3):A143–A4.
Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–84.
Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108(3):479.
X-x S, Yu Q. Intra-tumor heterogeneity of cancer cells and its implications for cancer treatment. Acta Pharmacol Sin. 2015;36(10):1219.
Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2010;1805(1):105–17.
Murtaza M, Dawson S-J, Pogrebniak K, Rueda OM, Provenzano E, Grant J, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun. 2015;6:8760.
Malapelle U, de-Las-Casas CM, Rocco D, Garzon M, Pisapia P, Jordana-Ariza N, et al. Development of a gene panel for next-generation sequencing of clinically relevant mutations in cell-free DNA from cancer patients. Br J Cancer. 2017;116(6):802–10.
Bennett CW, Berchem G, Kim YJ, El-Khoury V, Cell-free DNA. Next-generation sequencing in the service of personalized medicine for lung cancer. Oncotarget. 2016;7(43):71013.
Cani AK, Hovelson DH, Demirci H, Johnson MW, Tomlins SA, Rao RC. Next generation sequencing of vitreoretinal lymphomas from small-volume intraocular liquid biopsies: new routes to targeted therapies. Oncotarget. 2017;8(5):7989–98.
Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531–48.
Gerdes L, Iwobi A, Busch U, Pecoraro S. Optimization of digital droplet polymerase chain reaction for quantification of genetically modified organisms. Biomolecular detection and quantification. 2016;7:9–20.
Jovelet C, Madic J, Remon J, Honoré A, Girard R, Rouleau E, et al. Crystal digital droplet PCR for detection and quantification of circulating EGFR sensitizing and resistance mutations in advanced non-small cell lung cancer. PLoS One. 2017;12(8):e0183319.
Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci. 2012;109(36):14508–13.
Shu Y, Wu X, Tong X, Wang X, Chang Z, Mao Y, et al. Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple Cancer types. Sci Rep. 2017;7.1:583. https://doi.org/10.1038/541598-017-00520-1.
Bratman SV, Newman AM, Alizadeh AA, Diehn M. Potential clinical utility of ultrasensitive circulating tumor DNA detection with CAPP-Seq. Taylor Francis; 2015.
Chaudhuri A, Lovejoy A, Chabon J, Newman A, Stehr H, Say C, et al. CAPP-Seq circulating tumor DNA analysis for early detection of tumor progression after definitive radiation therapy for lung Cancer. International journal of radiation oncology• biology. Physics. 2016;96(2):S41–S2.
Mandel P. Les acides nucleiques du plasma sanguin chez l'homme. CR Acad Sci Paris. 1948;142:241–3.
Komatsubara KM, Sacher AG. Circulating Tumor DNA as a Liquid Biopsy: Current Clinical Applications and Future Directions. Oncology (Williston Park, NY). 2017;31(8).
Leon S, Shapiro B, Sklaroff D, Yaros M, Free DNA. In the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646–50.
Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46(5):318–22.
Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch R-D, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61(4):1659–65.
Anker P, Stroun M, Maurice PA. Spontaneous extracellular synthesis of DNA released by human blood lymphocytes. Cancer Res. 1976;36(8):2832–9.
Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102(45):16368–73.
Mouliere F, Rosenfeld N. Circulating tumor-derived DNA is shorter than somatic DNA in plasma. Proc Natl Acad Sci. 2015;112(11):3178–9.
Allis CD, Jenuwein T, Reinberg D. Epigenetics: CSHL Press; 2007.
Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausió J. Characterization of the stability and folding of H2A. Z chromatin particles implications for transcriptional activation. J Biol Chem. 2001;276(45):41945–9.
Anderson J, Thåström A, Widom J. Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Mol Cell Biol. 2002;22(20):7147–57.
Jansen A, Verstrepen KJ. Nucleosome positioning in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2011;75(2):301–20.
Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164(1):57–68.
genomics WDC. A nucleosome footprint reveals the source of cfDNA. Nat Rev Genet. 2016;17(3):125.
Ma X, Zhu L, Wu X, Bao H, Wang X, Chang Z, et al. Cell-free DNA provides a good representation of the tumor genome despite its biased fragmentation patterns. PLoS One. 2017;12(1):e0169231.
Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer—a survey. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2007;1775(1):181–232.
Yong E. Cancer biomarkers: written in blood. Nature. 2014;511:524–6.
Heidary M, Auer M, Ulz P, Heitzer E, Petru E, Gasch C, et al. The dynamic range of circulating tumor DNA in metastatic breast cancer. Breast Cancer Res. 2014;16(4):421.
Guo N, Lou F, Ma Y, Li J, Yang B, Chen W, et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci Rep. 2016;6
Wei Z, Wang W, Zitan Shu XZ, Zhang Y. Correlation between circulating tumor DNA levels and response to tyrosine kinase inhibitors (TKI) treatment in non-small cell lung Cancer. Medical science monitor: international medical journal of experimental and clinical research. 2017;23:3627–34.
Sacher AG, Paweletz C, Dahlberg SE, Alden RS, O’Connell A, Feeney N, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA oncology. 2016;2(8):1014–22.
af Hällström TM, Puhka M, Kallioniemi O. Circulating tumor DNA in early-stage breast cancer: personalized biomarkers for occult metastatic disease and risk of relapse? EMBO Molecular Medicine. 2015;7(8):994–5.
Khatami F, Aghayan HR, Sanaei M, Heshmat R, Tavangar SM, Larijani B. The potential of circulating tumor cells in personalized management of breast cancer: a systematic review. Acta Medica Iranica. 2017;55(3):175–93.
Olsson E, Winter C, George A, Chen Y, Howlin J, Tang MHE, et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO molecular medicine. 2015;7(8):1034–47.
Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early-and late-stage human malignancies. Science translational medicine. 2014;6(224):224ra24-ra24.
Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med 2010;2(20):20ra14-20ra14.
Dawson S-J, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin S-F, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–209.
Murtaza M, Dawson S-J, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108.
Sidransky D. Nucleic acid-based methods for the detection of cancer. Science. 1997;278(5340):1054–8.
Heitzer E, Auer M, Ulz P, Geigl JB, Speicher MR. Circulating tumor cells and DNA as liquid biopsies. Genome medicine. 2013;5(8):73.
Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61(1):112–23.
Mohammadi-asl J, Larijani B, Khorgami Z, Tavangar SM, Haghpanah V, Kheirollahi M, et al. Qualitative and quantitative promoter hypermethylation patterns of the P16, TSHR, RASSF1A and RARβ2 genes in papillary thyroid carcinoma. Med Oncol. 2011;28(4):1123–8.
Garcia-Murillas I, Beanney M, Epstein M, Howarth K, Lawson A, Hrebien S, et al. Abstract 2743: comparison of enhanced tagged-amplicon sequencing and digital PCR for circulating tumor DNA analysis in advanced breast cancer. Cancer Res. 2017;77(13 Supplement):2743.
Tzanikou E, Markou A, Politaki E, Koytsodontis G, Psyrri A, Georgoulias V, et al. Abstract 1725: Detection of <em>ESR1</em> D538G mutation in circulating tumor cells (CTCs) and paired circulating tumor DNA (ctDNA) samples of breast cancer patients. Cancer Research. 2017;77(13 Supplement):1725-.
Schiavon G, Hrebien S, Garcia-Murillas I, Pearson A, Tarazona N, Lopez-Knowles E, et al. Abstract 926: ESR1 mutations evolve during the treatment of metastatic breast cancer, and detection in ctDNA predicts sensitivity to subsequent hormone therapy. Cancer Res. 2015;75(15 Supplement):926.
Turner NC, Jiang Y, O'Leary B, Hrebien S, Cristofanilli M, Andre F, et al. Efficacy of palbociclib plus fulvestrant (P+F) in patients (pts) with metastatic breast cancer (MBC) and ESR1 mutations (mus) in circulating tumor DNA (ctDNA). Journal of Clinical Oncology. 2016;34(15_suppl):512-.
Baird RD, Rossum AV, Oliveira M, Beelen K, Garcia-Corbacho J, Mandjes IAM, et al. POSEIDON trial phase 1b results: Safety and preliminary efficacy of the isoform selective PI3K inhibitor taselisib (GDC-0032) combined with tamoxifen in hormone receptor (HR) positive, HER2-negative metastatic breast cancer (MBC) patients (pts) - including response monitoring by plasma circulating tumor (ct) DNA. Journal of Clinical Oncology. 2016;34(15_suppl):2520-.
Baselga J, Im S-A, Iwata H, Clemons M, Ito Y, Awada A, et al. Abstract S6–01: <em>PIK3CA</em> status in circulating tumor DNA (ctDNA) predicts efficacy of buparlisib (BUP) plus fulvestrant (FULV) in postmenopausal women with endocrine-resistant HR+/HER2– advanced breast cancer (BC): First results from the randomized, phase III BELLE-2 trial. Cancer Res 2016;76(4 upplement):S6–01-S6-.
Bosch A, Li Z, Bergamaschi A, Ellis H, Toska E, Prat A, et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor–positive breast cancer. Science translational medicine. 2015;7(283):283ra51-ra51.
Oshiro C, Kagara N, Naoi Y, Shimoda M, Shimomura A, Maruyama N, et al. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res Treat. 2015;150(2):299–307.
Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7(302):302ra133-302ra133.
Ma C, Bose R, Gao F, Freedman R, Telli M, Kimmick G, et al. Abstract CT011: Circulating tumor DNA (ctDNA) sequencing for <em>HER2</em> mutation (<em>HER2</em><sup>mut</sup>) screening and response monitoring to neratinib in metastatic breast cancer (MBC). Cancer Res 2017;77(13 Supplement):CT011-CT.
Qiu M, Wang J, Xu Y, Ding X, Li M, Jiang F, et al. Circulating tumor DNA is effective for the detection of EGFR mutation in non–small cell lung Cancer: a meta-analysis. Cancer Epidemiology Biomarkers & Prevention. 2014.
Goto T, Hirotsu Y, Amemiya K, Nakagomi T, Shikata D, Yokoyama Y, et al. Distribution of circulating tumor DNA in lung cancer: analysis of the primary lung and bone marrow along with the pulmonary venous and peripheral blood. Oncotarget. 2017;8(35):59268–81.
Hirsch FR, Varella-Garcia M, Bunn Jr PA, Franklin WA, Dziadziuszko R, Thatcher N, et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non–small-cell lung cancer. J Clin Oncol. 2006;24(31):5034–42.
Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169.
Bai H, Wang Z, Chen K, Zhao J, Lee JJ, Wang S, et al. Influence of chemotherapy on EGFR mutation status among patients with non–small-cell lung cancer. J Clin Oncol. 2012;30(25):3077–83.
Overman MJ, Modak J, Kopetz S, Murthy R, Yao JC, Hicks ME, et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed? J Clin Oncol. 2012;31(1):17–22.
Normanno N, Denis MG, Thress KS, Ratcliffe M, Reck M. Guide to detecting epidermal growth factor receptor (EGFR) mutations in ctDNA of patients with advanced non-small-cell lung cancer. Oncotarget. 2017;8(7):12501–16.
Henick BS, Goldberg SB, Narayan A, Rossi C, Rodney S, Kole AJ, et al. Circulating tumor DNA (ctDNA) to monitor treatment response and progression in patients treated with tyrosine kinase inhibitors (TKIs) and immunotherapy for EGFR-mutant non-small cell lung cancer (NSCLC). Proc Am Soc Clin Oncol. 2017;
De Mattos-Arruda L, Cortes J, Santarpia L, Vivancos A, Tabernero J, Reis-Filho JS, et al. Circulating tumour cells and cell-free DNA as tools for managing breast cancer. Nat Rev Clin Oncol. 2013;10(7):377–89.
Wang S, Han X. Hu X, Wang X, Zhao L, tang L, et al. clinical significance of pretreatment plasma biomarkers in advanced non-small cell lung cancer patients. Clin Chim Acta. 2014;430:63–70.
Jing C-W, Wang Z, Cao H-X, Ma R, Wu J-Z. High resolution melting analysis for epidermal growth factor receptor mutations in formalin-fixed paraffin-embedded tissue and plasma free DNA from non-small cell lung cancer patients. Asian Pac J Cancer Prev. 2013;14(11):6619–23.
Zhang H, Liu D, Li S, Zheng Y, Yang X, Li X, et al. Comparison of EGFR signaling pathway somatic DNA mutations derived from peripheral blood and corresponding tumor tissue of patients with advanced non-small-cell lung cancer using liquidchip technology. The Journal of Molecular Diagnostics. 2013;15(6):819–26.
Yeung KT, More S, Woodward B, Velculescu V, Husain H. Circulating tumor DNA for mutation detection and identification of mechanisms of resistance in non-small cell lung Cancer. Molecular Diagnosis & Therapy. 2017;21(4):375–84.
Audibert C, Shea M, Glass D, Kozak M, Caze A, Hohman R, et al. Use of FDA-approved vs. lab-developed tests in advanced non-small cell lung cancer. Proc Am Soc Clin Oncol. 2016;e20532. https://doi.org/10.1200/JCO.2016.34.15_suppl.e20532.
Nygaard AD, Garm Spindler K-L, Pallisgaard N, Andersen RF, Jakobsen A. The prognostic value of KRAS mutated plasma DNA in advanced non-small cell lung cancer. Lung Cancer. 2013;79(3):312–7.
Sherwood JL, Corcoran C, Brown H, Sharpe AD, Musilova M, Kohlmann A. Optimised pre-analytical methods improve KRAS mutation detection in circulating tumour DNA (ctDNA) from patients with non-small cell lung cancer (NSCLC). PLoS One. 2016;11(2):e0150197.
Paweletz CP, Oxnard GR, Feeney N, Hilton JF, Gandhi L, Do KT, et al. Abstract 3157: Serial droplet digital PCR (ddPCR) of plasma cell-free DNA (cfDNA) as pharmacodynamic (PD) biomarker in Phase 1 clinical trials for patients (pts) with KRAS mutant non-small cell lung cancer (NSCLC). Cancer Res. 2016;76(14 Supplement):3157-.
Bardelli A. Medical research: personalized test tracks cancer relapse. Nature. 2017;545(7655):417–8.
Geva S, Rozenblum AB, Ilouze M, Roisman L, Twito T, Dvir A, et al. P2.03b-047 the clinical impact of multiplex ctDNA Gene analysis in lung cancer. Journal of Thoracic Oncology. 12(1):S964.
Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7:11815.
Merriott DJ, Chaudhuri AA, Jin M, Chabon JJ, Newman A, Stehr H, et al. circulating tumor dna quantitation for early response assessment of immune checkpoint inhibitors for lung cancer. Int J Radiat Oncol Biol Phys. 99(2):S20–S1.
Khatami F, Larijani B, Heshmat R, Keshtkar A, Mohammadamoli M, Teimoori-Toolabi L, et al. Meta-analysis of promoter methylation in eight tumor-suppressor genes and its association with the risk of thyroid cancer. PLoS One. 2017;12(9):e0184892.
Cai X, Gao Y, Shen H, Laird P, Fan, J-B, Xu W, et al. Non-invasive diagnosis of early-stage lung cancer via targeted high-throughput DNA methylation sequencing of circulating tumor DNA (ctDNA). AACR; 2017.
Pantel K, Alix-Panabieres C. Liquid biopsy in 2016: circulating tumour cells and cell-free DNA in gastrointestinal cancer. Nat Rev Gastroenterol Hepatol. 2017;14(2):73–4.
Yan W, Zhang A, Powell MJ. Genetic alteration and mutation profiling of circulating cell-free tumor DNA (cfDNA) for diagnosis and targeted therapy of gastrointestinal stromal tumors. Chinese journal of cancer. 2016;35(1):68.
Sotoudeh M, Derakhshan MH, Abedi-Ardakani B, Nouraie M, Yazdanbod A, Tavangar SM, et al. Critical role of helicobacter pylori in the pattern of gastritis and carditis in residents of an area with high prevalence of gastric cardia cancer. Dig Dis Sci. 2008;53(1):27–33.
Malekzadeh R, Sotoudeh M, Derakhshan M, Mikaeli J, Yazdanbod A, Merat S, et al. Prevalence of gastric precancerous lesions in Ardabil, a high incidence province for gastric adenocarcinoma in the northwest of Iran. J Clin Pathol. 2004;57(1):37–42.
Szpechcinski A, Chorostowska-Wynimko J, Struniawski R, Kupis W, Rudzinski P, Langfort R, et al. Cell-free DNA levels in plasma of patients with non-small-cell lung cancer and inflammatory lung disease. Br J Cancer. 2015;113(3):476.
Shapiro B, Chakrabarty M, Cohn EM, Leon SA. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983;51(11):2116–20.
De Mattos-Arruda L, Olmos D, Tabernero J. Prognostic and predictive roles for circulating biomarkers in gastrointestinal cancer. Future Oncol. 2011;7(12):1385–97.
Howell JA, Khan SA, Knapp S, Thursz MR, Sharma R. The clinical role of circulating free tumor DNA in gastrointestinal malignancy. Transl Res. 2017;183(Supplement C):137–54.
Nasseri-Moghaddam S, Malekzadeh R, Sotoudeh M, Tavangar M, Azimi K, Sohrabpour AA, et al. Lower esophagus in dyspeptic Iranian patients: a prospective study. J Gastroenterol Hepatol. 2003;18(3):315–21.
Schwarzenbach H, Stoehlmacher J, Pantel K, Goekkurt E. Detection and monitoring of cell-free DNA in blood of patients with colorectal Cancer. Ann N Y Acad Sci. 2008;1137(1):190–6.
Boni L, Cassinotti E, Canziani M, Dionigi G, Rovera F, Dionigi R. Free circulating DNA as possible tumour marker in colorectal cancer. Surg Oncol. 2007;16:29–31.
Frattini M, Gallino G, Signoroni S, Balestra D, Battaglia L, Sozzi G, et al. Quantitative analysis of plasma DNA in colorectal cancer patients. Ann N Y Acad Sci. 2006;1075(1):185–90.
Frattini M, Gallino G, Signoroni S, Balestra D, Lusa L, Battaglia L, et al. Quantitative and qualitative characterization of plasma DNA identifies primary and recurrent colorectal cancer. Cancer Lett. 2008;263(2):170–81.
Danese E, Montagnana M, Minicozzi AM, De Matteis G, Scudo G, Salvagno GL, et al. Real-time polymerase chain reaction quantification of free DNA in serum of patients with polyps and colorectal cancers. Clin Chem Lab Med. 2010;48(11):1665–8.
Hedtke M, Haselmann V, Brechtel I, Duda A, Neumaier M. Use of liquid profiling/liquid biopsy to detect Ras mutations in cfdna of patients with metastatic colorectal cancer (mcrc). Clinical Chemistry and Laboratory Medicine. 2016;54(10):eA441.
Tavangar SM, Shariftabrizi A, Soroush AR. Her–2/neu over-expression correlates with more advanced disease in Iranian colorectal cancer patients. Med Sci Monit. 2005;11(3):CR123–CR6.
Pereira AAL, Morelli MP, Overman M, Kee B, Fogelman D, Vilar E, et al. Clinical utility of circulating cell-free DNA in advanced colorectal cancer. PLoS One. 2017;12(8):e0183949.
Spindler K-LG, Pallisgaard N, Vogelius I, Jakobsen A, Quantitative cell free DNA. KRAS and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clinical Cancer research. 2012:clincanres. 2011:0564.
Reinert T, Schøler LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65(4):625–34.
Lipson EJ, Velculescu VE, Pritchard TS, Sausen M, Pardoll DM, Topalian SL, et al. Circulating tumor DNA analysis as a real-time method for monitoring tumor burden in melanoma patients undergoing treatment with immune checkpoint blockade. Journal for immunotherapy of cancer. 2014;2(1):42.
Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu L, et al. The incidences and mortalities of major cancers in China, 2009. Chinese journal of cancer. 2013;32(3):106.
Bass AJ, Thorsson V, Shmulevich I, Reynolds SM, Miller M, Bernard B, et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202.
Zhang W. TCGA divides gastric cancer into four molecular subtypes: implications for individualized therapeutics. Chinese journal of cancer. 2014;33(10):469.
Nishida T, Biological HS. Clinical review of stromal tumors in the gastrointestinal tract. Histol Histopathol. 2000;15(4):1293–301.
Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer. 2011;11(12):865.
Lourenço N, Hélias-Rodzewicz Z, Bachet J-B, Brahimi-Adouane S, Jardin F, van Nhieu JT, et al. Copy-neutral loss of heterozygosity and chromosome gains and losses are frequent in gastrointestinal stromal tumors. Mol Cancer. 2014;13(1):246.
Astolfi A, Nannini M, Pantaleo MA, Di Battista M, Heinrich MC, Santini D, et al. A molecular portrait of gastrointestinal stromal tumors: an integrative analysis of gene expression profiling and high-resolution genomic copy number. Lab Investig. 2010;90(9):1285–94.
Nannini M, Astolfi A, Urbini M, Indio V, Santini D, Heinrich MC, et al. Integrated genomic study of quadruple-WT GIST (KIT/PDGFRA/SDH/RAS pathway wild-type GIST). BMC Cancer. 2014;14(1):685.
Gronchi A. Risk stratification models and mutational analysis: keys to optimising adjuvant therapy in patients with gastrointestinal stromal tumour. Eur J Cancer. 2013;49(4):884–92.
Şendur MAN, Özdemir NY, Akıncı MB, Uncu D, Zengin N, Aksoy S. Is exon mutation analysis needed for adjuvant treatment of gastrointestinal stromal tumor? World J Gastroenterol: WJG. 2013;19(1):144–6.
Wada N, Kurokawa Y, Takahashi T, Hamakawa T, Hirota S, Naka T, et al. Detecting secondary C-KIT mutations in the peripheral blood of patients with imatinib-resistant gastrointestinal stromal tumor. Oncology. 2016;90(2):112–7.
Boonstra PA, At E, Tibbesma M, Mathijssen RH, Atrafi F, Fv C, et al. Abstract 4951: dynamics of KIT exon 11 mutations in cell free plasma DNA of patients treated for advanced gastrointestinal stromal tumors: results from the Dutch GIST bio-databank. Cancer Res. 2017;77(13 Supplement):4951.
Maier J, Lange T, Kerle I, Specht K, Bruegel M, Wickenhauser C,et al. Detection of mutant free circulating tumor DNA in the plasma of patients with gastrointestinal stromal tumor harboring activating mutations of CKIT or PDGFRA. Clin Cancer Res. 2013;0765. https://doi.org/10.1158/1078-0432.CCR-13-0765.
Lan Y-T, Chen M-H, Fang W-L, Hsieh C-C, Lin C-H, Jhang F-Y, et al. Clinical relevance of cell-free DNA in gastrointestinal tract malignancy. Oncotarget. 2017;8(2):3009–17.
Fallahi P, Giannini R, Miccoli P, Antonelli A, Basolo F. Molecular diagnostics of fine needle aspiration for the presurgical screening of thyroid nodules. Current genomics. 2014;15(3):171–7.
Tavangar SM, Monajemzadeh M, Larijani B, Haghpanah V. Immunohistochemical study of oestrogen receptors in 351 human thyroid glands. Singap Med J. 2007;48(8):744–7.
Saffar H, Sanii S, Emami B, Heshmat R, Panah VH, Azimi S, et al. Evaluation of MMP2 and Caspase-3 expression in 107 cases of papillary thyroid carcinoma and its association with prognostic factors. Pathol Res Pract. 2013;209(3):195–9.
Sanii S, Tavangar SM. Cutaneous metastasis of medullary thyroid carcinoma as the initial manifestation of an otherwise limited malignancy: a case report. Am J Dermatopathol. 2011;33(7):716–8.
Haghpanah V, Abbas SI, Mahmoodzadeh H, Shojaei A, Soleimani A, Larijani B, et al. Paraplegia as initial presentation of follicular thyroid carcinoma. Journal of the College of Physicians and Surgeons--Pakistan : JCPSP. 2006;16(3):233–4.
Janku F, Huang HJ, Ramzanali NM, Hong DS, Karp DD. Fu S, et al. ultra-deep next-generation sequencing of plasma cell-free (cf) DNA from patients with advanced cancers. Proc Am Soc Clin Oncol. 2015;
Sandulache VC, Williams MD, Lai SY, Lu C, William WN, Busaidy NL, et al. Real-time genomic characterization utilizing circulating cell-free DNA in patients with anaplastic thyroid carcinoma. Thyroid. 2017;27(1):81–7.
Bible KC, Ryder M. Evolving molecularly targeted therapies for advanced-stage thyroid cancers. Nat Rev Clin Oncol. 2016;13(7):403–16.
Cote GJ, Evers C, Hu MI, Grubbs EG, Williams MD, Hai T, et al. Prognostic significance of circulating RET M918T mutated tumor DNA in patients with advanced medullary thyroid carcinoma. The Journal of Clinical Endocrinology & Metabolism. 2017;102(9):3591–9.
Brose MS, Cabanillas ME, Cohen EE, Wirth LJ, Riehl T, Yue H, et al. Vemurafenib in patients with BRAF V600E-positive metastatic or unresectable papillary thyroid cancer refractory to radioactive iodine: a non-randomised, multicentre, open-label, phase 2 trial. The Lancet Oncology. 2016;17(9):1272–82.
Khatami F, Larijani B, Tavangar SM. Circulating tumor BRAF mutation and personalized thyroid Cancer treatment. Asian Pacific journal of cancer prevention: APJCP. 2017;18(2):293–4.
Patel KB. Detection of circulating thyroid tumor DNA in patients with thyroid nodules. 2015.
Dakubo GD. Endocrine Cancer biomarkers in circulation. Cancer Biomarkers in Body Fluids: Springer; 2017. p. 457–80.
Pereira E, Camacho-Vanegas O, Anand S, Sebra R, Camacho SC, Garnar-Wortzel L, et al. Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers. PLoS One. 2015;10(12):e0145754.
Kajbafzadeh A-M, Payabvash S, Salmasi AH, Monajemzadeh M, Tavangar SM. Smooth muscle cell apoptosis and defective neural development in congenital ureteropelvic junction obstruction. J Urol. 2006;176(2):718–23.
Kamat AA, Sood AK, Dang D, Gershenson DM, Simpson JL, Bischoff FZ. Quantification of total plasma cell-free DNA in ovarian cancer using real-time PCR. Ann N Y Acad Sci. 2006;1075(1):230–4.
Harris FR, Kovtun IV, Smadbeck J, Multinu F, Jatoi A, Kosari F, et al. Quantification of somatic chromosomal rearrangements in circulating cell-free DNA from ovarian cancers. Sci Rep. 2016;6:29831.
Sarmadi S, Izadi-Mood N, Sotoudeh K, Tavangar SM. Altered PTEN expression; a diagnostic marker for differentiating normal, hyperplastic and neoplastic endometrium. Diagn Pathol. 2009;4(1):41.
Arend RC, Londono AI, Alvarez RD, Huh WK, Bevis KS, Leath CA, et al., editors. Circulating cell-free DNA: The future of personalized medicine in ovarian cancer management. Journal of Clinical Oncology; 2016: AMER SOC CLINICAL ONCOLOGY 2318 MILL ROAD, STE 800, ALEXANDRIA, VA 22314 USA.
Cristofanilli M, Turner NC, Bondarenko I, Ro J, Im SA, Masuda N, et al. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. The Lancet Oncology. 2016;17(4):425–39.
Acknowledgements
Special thanks to Endocrinology and Metabolism Research Center, Endocrinology and Metabolism Clinical Sciences Institute, Tehran University of Medical Sciences, Tehran, Iran.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author.
Funding
This article was a part of a larger project which was granted by the National Institute for Medical Research Development (NIMAD, Grant number: 957222).
Author information
Authors and Affiliations
Contributions
Professor Seyed Mohammad Tavangar made substantial contributions to conception and design, supervision, acquisition of data, and interpretation of data. Mrs. Fatemeh Khatami had been involved in drafting the manuscript or revising it critically for important intellectual content.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This manuscript does not report on or involve the use of any animal or human data or tissue, so ethical approval is not applicable in this section.
Consent for publication
This review article does not contain data from any individual person; consequently the consent for publication is “Not applicable” in this section.
Competing interests
All authors declare that they have no competing interests” in this section.
Rights and permissions
About this article
Cite this article
Khatami, F., Tavangar, S.M. Circulating tumor DNA (ctDNA) in the era of personalized cancer therapy. J Diabetes Metab Disord 17, 19–30 (2018). https://doi.org/10.1007/s40200-018-0334-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40200-018-0334-x