Abstract
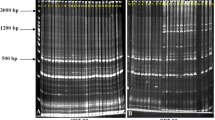
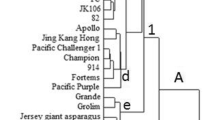
In this investigation, sixty morphologically different ginger rhizomes, collected from an active germplasm conservatory of High Altitude Research Station, Odisha, India, were characterized using morphological traits and molecular markers. Larger variations in morphological characters were observed among the 45 ginger accessions out of 60 accessions. Out of 22 markers, 9 ISSR and 13 SSR markers used for analysis generated 42 and 160 polymorphic bands accounting to 55% and 63.29% of the total polymorphic bands, respectively; this depicts that SSR markers have better efficiency over ISSR markers. A high degree of variation is detected in multivariate analysis (PCA and UPGMA) of both morphological traits and combined molecular markers, in which the ginger accessions clustered into five and four distinct groups. This explains the existence of large variation in morphological traits despite markers used. Genetic distance between the accessions determined from Nei’s dissimilarity matrix was used to find out the potential duplicates within the accessions collected. Twenty-nine accessions having mean genetic distance (MGD) less than threshold MGD (average MGD) value 0.174 were considered as ‘potential duplicate’ accessions and were reduced to one group, and the rest 31 were considered distinct genotype out of 60 accessions. From this analysis, 48.3% of the collection were considered as potential duplicates and deemed to be redundant. However, careful elimination of redundant through molecular analysis supported with passport data will help to eradicate existing synonyms used by farmers and improve management practices of the ginger germplasm conservatory.





Similar content being viewed by others
References
FAO (2009) Report on the state of the world’s plant genetic resources for food and agriculture. Food and Agriculture Organization of the United Nations, Rome
Van Hintum TJL, Boukema IW, Visser DL (1996) Reduction of duplication in a Brassica oleracea germplasm collection. Genet Resour Crop Evol 43:343–349
Rao NK (2004) Plant genetic resources: advancing conservation and use through biotechnology. Afr J Biotechnol 31:36–145
Hodgkin T, Brown AHD, Hintum TJLV, Morales EA (1995) Core collections of plant genetic resources. Wiley, Chichester
Phippen WB, Kresovich S, Candelas FG, McFerson JR (1997) Molecular characterization can quantify and partition variation among genebank holdings: a case study with phenotypically similar accessions of Brassica oleracea var. Capitata L. (cabbage) “Golden Acre”. Theor Appl Genet 94:227–234
Zhang D, Ghislain M, Huaman Z, Rodriguez F, Cervantes J (1997) Identifying duplicates in sweet potato germplasm using RAPD, International Potato centre, Programme report 1995–1996. Lima, Peru, pp 90–96
Treuren R, Van Soest LJM, Van Hintum TJL (2001) Marker-assisted rationalisation of genetic resource collections: a case study in flax using AFLPs. Theor Appl Genet 103:144–152
Chavarriaga-Aguirre P, Maya MM, Tohme J, Duque MC, Iglesias C, Bonierbale MW, Kresovich S, Kochert G (1999) Using microsatellites, isozymes and AFLPs to evaluate genetic diversity and redundancy in the cassava core collection and to assess the usefulness of DNA-based markers to maintain germplasm collections. Mol Breed 5:263–273
Subudhi E, Das A, Joshi RK, Mohanty S, Nayak S (2016) Genetic diversity analysis and redundant identification in 48 core collections of Zingiber officinale Rosc. (Zingiberaceae). Bras Bot 39:869–883
HerbaZest (2018) Importance of Ginger. https://www.herbazest.com/herbs/ginger/importance-of-ginger. Accessed 10 Jan 2018
Hassa MF, Hussein S, Senosi YE, Mansour MK, Amin A (2018) Role of ginger as anti-inflammatory and anti-apoptotic in protection of liver damage induced by metalaxyl fungicide in male albino rats. J Clin Exp Pathol 8:1–8
Mashhadi NS, Ghiasvand R, Askari G, Hariri M, Darvishi L, Mofid MR (2013) Anti-oxidative and anti-inflammatory effects of ginger in health and physical activity: review of current evidence. Int J Prev Med 4:36–42
Liu Y, Whelan RJ, Pattnaik BR, Ludwig K, Subudhi E, Rowland H, Claussen N, Zucker N, Uppal S, Kushner DM, Felder M, Patankar MS, Kapur A (2012) Terpenoids from Zingiber officinale (Ginger) induce apoptosis in endometrial cancer cells through the activation of p53. PLoS ONE 7:e53178
Engels JMM, Visser L (2003) A guide to effective management of germplasm collections. IPGRI handbooks for genebanks No. 6. International Plant Genetic Resources Institute
Mohanty DC, Panda BS (1994) Genetic resources of Ginger. In: Chadha KL, Rethinam P (eds) Advances in horticulture, plantation and spices crops, Part-1, vol 9. Malhotra Publishing House, New Delhi, pp 151–167
Motilal LA, Zhang D, Mischke SL, Meinhardt W, Umaharan P (2013) Microsatellite-aided detection of genetic redundancy improves management of the International Cocoa Genebank Trinidad. Tree Genet Genom 9:1395–1411
Fu YB (2006) Redundancy and distinctness in flax germplasm as revealed by RAPD dissimilarity. Plant Genet Resour 4:117–124
Song ZP, Xu X, Wang B, Chen JK, Lu BR (2003) Genetic diversity in the northern most Oryza rufipogon populations estimated by SSR markers. Theor Appl Genet 107:1492–1499
Das A, Gaur M, Barik DP, Subudhi E (2017) Genetic diversity analysis of 60 ginger germplasm core accessions using ISSR and SSR markers. Plant Biosyst 151(5):522–532
Gherardi M, Mangin B, Goffinet B, Bonnet D, Huguet T (1998) A method to measure genetic distance between all ogamous populations of alfalfa (Medicago sativa) using RAPD molecular marker. Theor Appl Genet 98:406–412
Prevost A, Wilkins MJ (1999) new system for comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:661–668
Roberts DW (2015) labdsv: Ordination and multivariate analysis for ecology. R package version, 2007 journal name in full R topic documented. R Top Doc. pp 59
Suzuki R, Shimodaira H (2006) Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542
Rohlf FJ (2000) NTSYS-pc: Numerical taxonomy and multivariate analysis system (Version 2.1). Exeter Software, New York
Ravindran PN, Babu NK (eds) (2005) Introduction. In: Ginger-the genus zingiber. CRC Press, Boca Raton, pp 15–85
Singh PP, Singh VB, Singh A, Singh HB (1999) Evaluation of different ginger cultivars for growth, yield and quality character under Nagaland condition. J Med Aromat Plant Sci 21:716–718
Pourhosseini SH, Hadian J, Sonboli A, Nejad Ebrahimi S, Mirjalili MH (2018) Genetic and chemical diversity in Perovskia abrotanoides Kar. (Lamiaceae) populations based on ISSRs markers and essential oils profil. Chem Biodivers 15:e1700508
Cooper HD, Spillane C, Hodgkin T (2001) Broadening the genetic base of crops: an overview. In: Spillane C, Hodgkin T (eds) Broadening the Genetic Base of Crop Production. CABI Publishing, Wallingford, pp 1–23
Jatoi SA, Kikuchi A, Mimura M, San-San-Yi WK (2008) Relationships of Zingiber species, and genetic variability assessment in ginger (Zingiber officinale) accessions from ex situ genebank, on-farm and rural markets. Breed Sci 58:261–270
Mahar KS, Rana TS, Ranade S (2012) Molecular analyses of genetic variability in soap nut (Sapindus mukorossi Gaertn.). Ind Crops Prod 34:1111–1118
Prem J, Kizhakkayil J, Thomas E, Dhanya K, Syamkumar S, Sasikumar B (2008) Molecular characterization of primitive, elite and exotic Ginger genotypes to protect the biowealth of elite Ginger accessions. J Spices Arom Crop 17:85–90
Senan S, Kizhakayil D, Sheeja TE, Sasikumar B, Bhat AI, Parthasarathy VA (2013) Novel polymorphic microsatellite markers from turmeric, Curcuma longa L. (Zingiberaceae). Acta Botanica Croatica 72:407–412
Ganie SH, Ali Z, Das S, Srivastava PS, Sharma MP (2015) Genetic diversity and chemical profiling of different populations of Convolvulus pluricaulis (convolvulaceae): an important herb of ayurvedic medicine. 3 Biotech 5:295–302
Ravishanker KS, Baranwal DK, Chatterjee A, Solankey SS (2013) Genetic diversity based on cluster and principal component analyses for yield and quality attributes in ginger (Zingiber officinale Rosc.). Int J Plant Breed Genet 7:159–168
Hailegiorgis D, Mesfin M, Genet T (2011) Genetic divergence analysis on some bread wheat genotypes grown in Ethiopia. J Central Eur Agric 12:344–352
Pandotra P, Gupta AP, Husain MK, Gandhiram GS (2013) Evaluation of genetic diversity and chemical profile of ginger cultivars in north-western Himalayas. Biochem Syst Ecol 48:281–287
Lund B, Ortiz R, Skovgaard IM, Waugh R (2003) Andersen SB analysis of potential duplicates in barley gene bank collections using re-sampling of microsatellite data. Theor Appl Genet 106:1129–1138
Senula A, Keller ERJ (2000) Morphological characterization of a garlic core collection and establishment of a virus-free in vitro gene bank. Allium Improv Newsl 10:3–5
Acknowledgement
The authors are grateful to the President and Dean for providing valuable infrastructure at Centre of Biotechnology, Siskha ‘O’ Anusandhan (Deemed to be University), Bhubaneswar, and Mr. P. Sial, in charge of HARS, Pottangi, for providing ginger germplasm.
Funding
Funding for this study was received from the Department of Science and Technology, New Delhi, Grant No. SERB/SR/SO/PS/41/2012.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to publish this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance Statement
This manuscript deals with morphological and genetic variability of the ginger accessions collected from gene bank of Odisha. The phenotypic and genotypic correlation is observed within accessions. The detection and elimination of duplicate ginger germplasm through combined ISSR and SSR marker analysis can help to improve the management practices of the ginger germplasm conservatory of Odisha.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Das, A., Sahoo, R.K., Barik, D.P. et al. Identification of Duplicates in Ginger Germplasm Collection from Odisha Using Morphological and Molecular Characterization. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 90, 1057–1066 (2020). https://doi.org/10.1007/s40011-020-01178-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-020-01178-y