Abstract
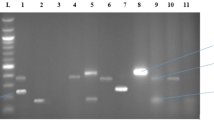
Antibiotic resistance genes are considered to be emerging contaminants. Considering the limited number of papers on detection of antibiotic resistance genes in metal-polluted soils of Iran and due to the fact that nonclinical strains carrying resistance determinants can be the origin of resistance genes in clinical isolates, the present study was conducted to detect some resistance genes (i.e., blaTEM, vanA, tetB, strA, and aac(3)-II) in the most prevalent culturable bacteria isolated from soils around mines. A total of 70 species of Pseudomonas, Azotobacter, Enterobacter, and Bacillus were isolated from soils under different land uses. Polymerase chain reaction was used to detect resistance genes in the isolates. The blaTEM gene was the most abundant gene detected in the isolates (45.71%). The number of Azotobacter, Pseudomonas, and Enterobacter isolates containing blaTEM was higher in the agricultural and pasture soils than in the mining waste soils (28.57%, 57.14%, and 20%, respectively), but the pasture and mining waste soils proved to harbor more Bacillus species containing blaTEM compared to the agricultural soils (64.28%, 50%, and 42.86%, respectively). The vanA gene was found in 5.71% of all the strains, and only one Pseudomonas isolates harbored aac(3)-II. The tetB and strA genes were not detected in any of the isolates. More than 77% of the isolates were phenotypically resistant to β-lactams, and 28.57%, 40%, and 31.43% of them were resistant to streptomycin, vancomycin, and tetracyclines, respectively. Overall, the high number of bacteria containing at least one resistance gene isolated from the samples indicated the persistence of environmental reservoirs of resistance genes in the metal-polluted soils.




Similar content being viewed by others
References
Adarsh V, Mishra M, Chowdhury S, Sudarshan M, Thakur A, Chaudhuri SR (2007) Studies on metal microbe interaction of three bacterial isolates from East Calcutta Wetland. J Biol Sci 7:80–88
Aendekerk S, Diggle SP, Song Z, Høiby N, Cornelis P (2005) The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology 151:1113–1125
Alonso A, Morales G, Escalante R, Campanario E, Sastre L, Martinez JL (2004) Overexpression of the multidrug efflux pump SmeDEF impairs Stenotrophomonas maltophilia physiology. J Antimicrob Chemother 53:432–434
Arias CA, Murray BE (2015) A new antibiotic and the evolution of resistance. N Engl J Med 372:1168–1170
Avner BS, Fialho AM, Chakrabarty AM (2012) Overcoming drug resistance in multi-drug resistant cancers and microorganisms: a conceptual framework. Bioengineered 3:262–270
Baker-Austin C, Wright MS, Stepanauskas R, McArthur J (2006) Co-selection of antibiotic and metal resistance. Trends Microbiol 14:176–182
Bradford PA (2001) Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev 14:933–951
Capkin E, Terzi E, Altinok I (2015) Occurrence of antibiotic resistance genes in culturable bacteria isolated from Turkish trout farms and their local aquatic environment. Dis Aquat Org 114:127–137
Collins C, Lyne P (1987) Microbiological methods. Butter Morths and Co (Publishers) Ltd., London
D’Costa VM, Griffiths E, Wright GD (2007) Expanding the soil antibiotic resistome: exploring environmental diversity. Curr Opin Microbiol 10:481–489
Dantas G, Sommer MO (2012) Ecological and clinical consequences of antibiotic subsistence by environmental microbes. In: Dantas G, Sommer MOA (eds) Antimicrobial resistance in the environment, 1st edn. Wiley, Hoboken
De Gheldre Y, Avesani V, Berhin C, Delmée M, Glupczynski Y (2003) Evaluation of Oxoid combination discs for detection of extended-spectrum β-lactamases. J Antimicrob Chemother 52:591–597
Dubois V, Arpin C, Noury P, Quentin C (2002) Clinical strain of Pseudomonas aeruginosa carrying a blaTEM-21 gene located on a chromosomal interrupted TnA type transposon. Antimicrob Agents Chemother 46:3624–3626
El-Enbaawy MI, Yousif AA (2006) β-Lactamase gene in multi-drug resistant clinical bacterial isolates from Egyptian food animal species. Arab J Biotechnol 9:71–82
Faldynova M, Videnska P, Havlickova H, Sisak F, Juricova H, Babak V, Steinhauser L, Rychlik I (2013) Prevalence of antibiotic resistance genes in faecal samples from cattle, pigs and poultry. Vet Med Czech 58:298–304
Graham DW, Knapp CW, Christensen BT, McCluskey S, Dolfing J (2016) Appearance of β-lactam resistance genes in agricultural soils and clinical isolates over the 20th century. Sci Rep 6:21550
Henriques IS, ALvEs A, Saavedra MJ, Montforts MH, Correia A (2011) Environmental antibiotic resistome: new insights from culture-independent approaches. In: Keen PL, Montforts MHMM (eds) Antimicrobial resistance in the environment. Wiley, Hoboken, pp 123–148
Hu J, Zhao J, Wang D, Li X, Zhang D (2018) Effect of diclofenac on the production of volatile fatty acids from anaerobic fermentation of waste activated sludge. Bioresour Technol 254:7–15
Iredell J, Brown J, Tagg K (2016) Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. BMJ 352:h6420
Jensen H (1942) Nitrogen fixation in leguminous plants. I. General characters of root-nodule bacteria isolated from species of Medicago and Trifolium in Australia. Proc Linn Soc NSW 67:205–212
Keen PL, Montforts MH (2012) Antimicrobial resistance in the environment. Wiley, Hoboken, pp 251–265
Knapp CW, Dolfing J, Ehlert PA, Graham DW (2009) Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ Sci Technol 44:580–587
Knapp CW, McCluskey SM, Singh BK, Campbell CD, Hudson G, Graham DW (2011) Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived Scottish soils. PLoS ONE 6:e27300
Knapp CW, Callan AC, Aitken B, Shearn R, Koenders A, Hinwood A (2017) Relationship between antibiotic resistance genes and metals in residential soil samples from Western Australia. Environ Sci Pollut Res 24:2484–2494
Krumperman PH (1983) Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 46:165–170
Lefkowitz JR, Duran M (2009) Changes in antibiotic resistance patterns of Escherichia coli during domestic wastewater treatment. Water Environ Res 81:878–885
Malik A, Aleem A (2011) Incidence of metal and antibiotic resistance in Pseudomonas spp. from the river water, agricultural soil irrigated with wastewater and groundwater. Environ Monit Assess 178:293–308
Martinez JL (2009) Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut 157:2893–2902
Máthé I, Benedek T, Táncsics A, Palatinszky M, Lányi S, Márialigeti K (2012) Diversity, activity, antibiotic and heavy metal resistance of bacteria from petroleum hydrocarbon contaminated soils located in Harghita County (Romania). Int Biodeterior Biodegrad 73:41–49
McKinney CW, Loftin KA, Meyer MT, Davis JG, Pruden A (2010) Tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence. Environ Sci Technol 44:6102–6109
Muller JF, Stevens AM, Craig J, Love NG (2007) Transcriptome analysis reveals that multidrug efflux genes are upregulated to protect Pseudomonas aeruginosa from pentachlorophenol stress. Appl Environ Microbiol 73:4550–4558
Patel J, Cockerill F, Alder J, Bradford P, Eliopoulos G, Hardy D (2014) Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI Stand Antimicrob Susceptibility Test 34:1–226
Popowska M, Rzeczycka M, Miernik A, Krawczyk-Balska A, Walsh F, Duffy B (2012) Influence of soil use on prevalence of tetracycline, streptomycin, and erythromycin resistance and associated resistance genes. Antimicrob Agents Chemother 56:1434–1443
Pruden A, Arabi M (2012) Quantifying anthropogenic impacts on environmental reservoirs of antibiotic resistance. In: Keen PL, Montforts MHMM (eds) Antimicrobial resistance in the environment. Wiley, Hoboken, pp 173–201
Ram S, Vajpayee P, Shanker R (2007) Prevalence of multi-antimicrobial-agent resistant, shiga toxin and enterotoxin producing Escherichia coli in surface waters of river Ganga. Environ Sci Technol 41:7383–7388
Ramos JL, Duque E, Gallegos M-T, Godoy P, Ramos-Gonzalez MI (2002) Mechanisms of solvent tolerance in gram-negative bacteria. Ann Rev Microbiol 56:743–768
Ren Q, Paulsen IT (2005) Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput Biol 1:e27
Roberts MC (2012) Mechanisms of bacterial antibiotic resistance and lessons learned from environmental tetracycline resistant bacteria. In: Keen PL, Montforts MHMM (eds) Antimicrobial resistance in the environment. Wiley, New York, pp 93–121
Roberts MC (2017) Antibiotic-resistant environmental bacteria and their role as reservoirs in disease. In: Hurst C (ed) Modeling the transmission and prevention of infectious disease. Springer, Berlin, pp 187–212
Sinegani AAS, Younessi N (2017) Antibiotic resistance of bacteria isolated from heavy metal-polluted soils with different land uses. J Glob Antimicrob Resist 10:247–255
Soge O, Tivoli L, Meschke J, Roberts M (2009) A conjugative macrolide resistance gene, mef (A), in environmental Clostridium perfringens carrying multiple macrolide and/or tetracycline resistance genes. J Appl Microbiol 106:34–40
Stepanauskas R, Glenn TC, Jagoe CH, Tuckfield RC, Lindell AH (2006) Coselection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ Microbiol 8:1510–1514
Tendencia EA, de la Peña LD (2001) Antibiotic resistance of bacteria from shrimp ponds. Aquaculture 195:193–204
Thorburn AL (1983) Paul Ehrlich: pioneer of chemotherapy and cure by arsenic (1854–1915). Sex Transm Inf 59:404–405
Upadhyay S, Kumar N, Singh V (2013) Antibiotic resistance and pathogen inhibition by Azotobacter isolates. Pantnagar J Res 11:254–256
Vliegenthart J, Ketelaar-van Gaalen P, van de Klundert J (1989) Nucleotide sequence of the aacC2 gene, a gentamicin resistance determinant involved in a hospital epidemic of multiply resistant members of the family Enterobacteriaceae. Antimicrob Agents Chemother 33:1153–1159
Volkmann H, Schwartz T, Bischoff P, Kirchen S, Obst U (2004) Detection of clinically relevant antibiotic-resistance genes in municipal wastewater using real-time PCR (TaqMan). J Microbiol Methods 56:277–286
Wang Y, Wang D, Liu Y, Wang Q, Chen F (2017) Triclocarban enhances short-chain fatty acids production from anaerobic fermentation of waste activated sludge. Water Res 127:150–161
Acknowledgements
This research was supported by Bu-Ali Sina University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editorial responsibility: M. Abbaspour.
Rights and permissions
About this article
Cite this article
Younessi, N., Safari Sinegani, A.A. & Khodakaramian, G. Detection of antibiotic resistance genes in culturable bacteria isolated from soils around mines in Hamedan, Iran. Int. J. Environ. Sci. Technol. 16, 7643–7652 (2019). https://doi.org/10.1007/s13762-018-02178-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-018-02178-2