Abstract
Euglossini bees are able to fly long distances, which could help to maintain gene flow among widely separated populations. In order to investigate if different environmental conditions affect morphological variation in Euglossa annectans and Euglossa truncata, we analyzed the patterns of venation of the forewings of 310 individuals, sampled in the same six locations for the two species. Populations from the two species clustered in a similar way, following the phytophysiognomy of the sampling sites. These populations also presented little or no population structure. Based on our results, we suggest that the forest fragmentation is not a problem for these species. The tendency of samples to group based on site phytophysiognomy can be explained by phenotypic plasticity or local adaptations.
Similar content being viewed by others
1 Introduction
Among the 20,000 bee species worldwide (Michener 2000), the Euglossini are especially relevant due to their representability in the Neotropical region, where they compose up to 25% of the total bee community in some areas (Roubik and Hanson 2004). They are exclusive pollinators of almost 700 species of Neotropical Orchidaceae (Ramírez et al. 2002; Oliveira and Nemésio 2003; Roubik and Hanson 2004; Oliveira 2006) and many other plant species, including Araceae (Williams and Dressler 1976), Euphorbiaceae (Armbruster and Webster 1979), Haemodoraceae (Buchmann 1981), Apocynaceae, Loganiaceae, and Malvaceae (Ackerman 1983), Solanaceae (Soares et al. 1989), Pentaphylacaceae (Melo 1995), Lecythidaceae (Knudsen and Mod 1996), Clusiaceae (Nogueira et al. 1998), Bromeliaceae (Siqueira-Filho and Machado 2008), Annonaceae (Teichert et al. 2009), and Plantaginaceae (Cappellari et al. 2009). These bees are restricted to the Neotropical region, and their geographic distribution ranges between 29 °N and 32 °S parallels. Euglossini males are known for their great flight capacity; some reports mention that males can fly more than 20 Km per day in a search for nectar and floral fragrances, in a non-fragmented environment (Janzen 1971; Kroodsma 1975; Ackerman et al. 1982 and Eltz et al. 2003). They fly long distances searching for aromatic compounds from multiple floral and non-floral sources, subsequently used during courtship display (Dressler 1982; Eltz et al. 2005). However, in spite of the great flight range of the males, forest fragmentation could be a barrier for these bees (Tonhasca et al. 2003; Brosi 2009), making them particularly susceptible to deforestation (Roubik and Hanson 2004; Cerântola et al. 2011). According to Zayed (2009), habitat fragmentation is a direct result of habitat loss and could affect the survival of populations through genetic isolation and subsequent inbreeding.
Due to the importance of Euglossini bees as pollinators and the concerns about the impacts of forest fragmentation, we examined morphological variability variation in two species, collected from forest fragments in Southeast Brazil. We employed geometric morphometrics analysis of wings, an effective methodology for the discrimination of cryptic species (Francisco et al. 2008, Hurtado-Burillo et al. 2016), geographic ecotypes (Francoy et al. 2011, Bonatti et al. 2014) and for the identification of species (Francoy et al. 2009, 2012) and subspecies (Oleksa and Tofilski 2014; Silva et al. 2015). Though wing shape is relatively conserved among individuals of same species, this technique has shown to be a sensitive tool for very fine differentiation among groups, (Debat et al. 2003; De Souza et al. 2015). This work also aims to provide information that could assist in the development of conservation strategies for the Euglossini.
2 Materials and methods
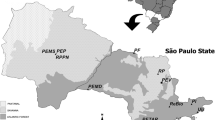
Euglossini males (126 Euglossa annectans and 184 Euglossa truncata) were collected from six different forest fragments in São Paulo state, Brazil, according to the methodology proposed by Rebêlo and Garófalo (1997) and identified in the laboratory (Figure 1; Table I) (The codes used for the sampled areas are as follows: APASJ—Área de Proteção Ambiental Serra do Japi; EEC—Estação Ecológica dos Caetetus; EEJ—Estação Ecológica do Jataí; EERP—Estação Ecológica de Ribeirão Preto; MV—Mata da Virgínia; PFBJ—Parque Estadual das Furnas do Bom Jesus). All the bees used in this work are deposited in the Collection of Bees and Solitary Wasps (Coleção de Abelhas e Vespas Solitárias—CAVES) of the Department of Biology of the Faculty of Philosophy, Science and Letters in Ribeirão Preto—University of São Paulo, Brazil.
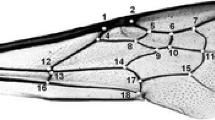
For the morphometric analyses, the right forewings were carefully cut from the bodies of the bees using an entomological scissors and placed on microscope slides and sealed with a cover slip. We photographed the 310 forewings using a digital camera attached to a stereoscopic microscope, and the images were stored as digital files. Eighteen homologous landmarks were plotted on each wing, consisting of one landmark per vein intersection (Figure 2) using tpsDig2 version 2.12 (Rohlf 2008). The Cartesian coordinates of each landmark were recorded and used as input data into MorphoJ 1.05f (Klingenberg 2011) to perform the morphometric analysis. The images were Procrustes aligned and the data were analyzed using canonical variate analysis (CVA) and discriminant function analysis (DFA). All software used in this work is freely available on the following internet websites: http://life.bio.sunysb.edu/morph/ and http://www.flywings.org.uk/morphoj_page.htm.
We also used MorphoJ (Klingenberg 2011) to calculate the Procrustes distances among the groups and to perform the cross-validation test on the discriminant analysis; bees were grouped according to their sampling locations. The Mahalanobis square distance and Procrustes distances between mean shapes of the groups were extracted and used in the construction of dendrograms of morphological proximity (based on neighbor-joining distances) using MEGA 5.0 (Tamura et al. 2011). We ran a Mantel test using TFPGA software (Miller 1997) on the Mahalanobis square distances between the centroids of the groups and the geographic distances between the sites (measured by Google Earth 7.1.5.1557/2015). We also employed the Mantel test on the Procrustes distances and the geographic distances between sites.
3 Results
3.1 Euglossa annectans
Based on geometric morphometric analysis of the Eg. annectans samples, canonical variation analysis explained 60.04% of the variance on the first two axes (CVA1 explained 34.25% and CVA2 explained 25.78%) (Supp. Figure S1). The plot of the individuals on the first two CVA axes showed some degree of superposition of the individuals of all samples, with a tendency to separate them. Based on the first CVA axes, we find two groups, where one group is placed in the right-hand quadrant, with EEC, MV, and APASJ, and the other group in the left-hand quadrant with PFBJ, EEJ, and EERP.
The CVA indicated that all groups formed clusters significantly different from each other (P < 0.05), except for MV and APASJ (P = 0.06). The Mahalanobis distance varied from 2.41 (between APASJ x EEC) to 3.93 (between EERP x PFBJ) and the Procrustes distances varied from 0.009 (between APASJ x EERP and APASJ x MV) to 0.016 (between EERP x PFBJ) (Table II).
The two dendrograms of morphological proximity presented a similar topology (Figure 3), indicating similar relationships between populations. MV and APASJ are placed together in one branch, close to the populations of EEC and EERP. More distant to these branches, the populations of EEJ and PFBJ are placed in another branch. The only difference between the two dendograms is EEC placed together with EERP on the dendogram based on the Procrustes distances, while they are separated in the other dendogram.
Neighbor-joining dendrogram of Euglossa annectans groups constructed based on the Mahalanobis square distances between the centroids (a) and Procrustes distance (b). See legend to Figure 1 for abbreviations.
The mean classification of discriminant analysis and cross-validation test were 93.4 and 58.3%, respectively (Table III). The Mantel test correlating the geographic distances between fragments and the Mahalanobis square distance and Procrustes distance between groups presented non-significant results (r = − .0706; P = 0.57 and r = .0688; P = 0.55, respectively).
3.2 Euglossa truncata
The first two axes of the canonical variation analysis explained 70.64% of the variance (CVA1 explained 43.22% and CVA2 explained 27.42%) (Supp. Figure S2). The CVA indicated that all groups formed clusters significantly different from each other (P < 0.05), except for the groups from EERP and PFBJ (P = 0.06). The scatterplot shows a clear tendency of separation of all samples, except for EEC and EERP.
The Mahalanobis distance varied from 1.78 (between APASJ x EEC) to 4.91 (between EERP x PFBJ) and the Procrustes distances among them varied from 0.008 (between EEC x EEJ and EEJ x PFBJ) to 0.024 (between EERP x MV) (Table IV).
The dendrogram of morphological proximity, built based on the Mahalanobis square distances between the centroids of the groups (Figure 4a), placed the bees grouped on the top, on the right side, one branch with APASJ alone and on the left side, and another with EEC and EERP together. On the bottom, on the right side, there is a branch with PFBJ alone and on the left side, another branch with EEJ and MV. We built a dendrogram based on Procrustes distance (Figure 4b), and the results corroborate with the Mahalanobis distance dendrogram with only one exception: the inverted positions of EEJ and PFBJ.
Neighbor-joining dendrogram of Euglossa truncata groups constructed based on the Mahalanobis square distances between the centroids (a) and Procrustes distance (b). See legend to Figure 1 for abbreviations.
The mean classification of discriminant analysis and cross-validation test were 98 and 72%, respectively (Table V). The Mantel test correlating the geographic distances between fragments and the Mahalanobis square distance and Procrustes distance between groups presented non-significant results (r = 0.1976; P = 0.23 and r = .2638 Pl = 0.72) between the groups.
We also performed a CVA analysis with all samples from both species together (Figure 5), and based on Mahalanobis and Procrustes distances, we built a dendrogram of morphological similarity. The two dendrograms showed the same pattern of site grouping for the two species, and complete separation between them.
Neighbor-joining dendrogram of samples of Euglossa annectans (1) and Euglossa truncata (2) from different collection sites, constructed based on the Mahalanobis square distances between the centroids (a) and Procrustes distances (b). See legend to Figure 1 for abbreviations.
4 Discussion
Many publications have shown the power of geometric morphometric (GM) technique for recognition of cryptic species (Francisco et al. 2008) and geographic ecotypes (Francoy et al. 2011; Bonatti et al. 2014) as well as for identification of species (Francoy et al. 2009, 2012) and subspecies (Oleksa and Tofilski 2014; Silva et al. 2015) of bee populations. In spite of the similarity found among the populations (shown by CVA analysis), the GM technique was able to demonstrate the same pattern of grouping of samples for the two species, according to each collection site. All of our collection sites are located within Atlantic forest fragments, some of them in association with Savanah areas. This forest is one of the greatest hotspots of biodiversity in the world (Myers et al. 2000 and Carnaval et al. 2009), and in the last 200 years, its coverage has been reduced from 81.8 (Victor 1975) to 7% (MMA) as a result of anthropic activities (urbanization and agribusiness). Habitat fragmentation is one of the main concerns for decline of bee populations (Cane 2001; Zayed 2009). When populations decrease in size, genetic factors such as inbreeding depression and loss of genetic diversity (which can reduce adaptability) could accelerate the extinction process (Brook et al. 2002; Frankham 2003; Spielman et al. 2004 O’Grady et al. 2008).
Several studies have shown that some bee species have difficulty in crossing between fragmented areas. Milet-Pinheiro and Schlindwein (2005), in a study of males of 16 Euglossini species at various sites that ranged from the middle of an Atlantic forest fragment to 500 m away in a sugar cane plantation, observed that only two species (Eulaema nigrita and Euglossa cordata) came out of the forest and flew over the monoculture region. Another research done in the Amazon forest observed that the conversion of some forest areas into pasture has also created a barrier limiting the dispersion of Euglossini males (Powell and Powell 1987). However, studies using various types of markers, including mitochondrial DNA, microsatellites, and allozymes have shown that Euglossini bees are usually capable of flying long distances, exhibiting high genetic diversity and low population structure (Souza et al. 2010; Cerântola et al. 2011 and Zimmermann et al. 2011; Suni and Brosi 2012). Souza et al. (2010), in a study of diploid Euglossini males of in a Neotropical region, observed high gene flow and a large effective population size in a population of Euglossa cordata, since they found high genetic diversity, a low degree of population structure, and very few diploid males. In another study, Rocha-Filho et al. (2013) assessed gene variation and genetic differentiation among populations of four abundant Euglossini species sampled in two areas, Picinguaba (mainland) and Anchieta Island, Ubatuba, SP, southeastern Brazil. They found no significant genetic differentiation between the island and mainland samples of Euglossa cordata, Eulaema cingulata, and Eulaema nigrita; though significant gene differentiation between the samples of Euglossa stelfeldi was detected. These results indicated that the distance between the island and the mainland did not constitute a geographic barrier for these Euglossini bees. The absence of populational structuring of three of the four species corroborated previous reports on these bees, characterized by large populations, with high gene diversity and gene flow and very low frequencies of diploid males. Suni and Brosi (2012) showed that the mobility of Euglossini bees differs greatly among species in fragmented habitats and that some of these bees have high levels of gene flow compared to other bee groups. One similar point indicated in all the studies cited above is the fact that the number of migrating individuals is large enough to maintain gene flow between populations (Suni and Brosi 2012; Rocha-Filho et al. 2013). However, even larger species, with a high dispersal capacity, do not migrate in large numbers (Milet-Pinheiro and Schlindwein 2005). These two facts taken together can help explain the similarities and the differences found among populations. Although we worked with morphological data, our results for the two species demonstrated some degree of similarity between the groups, despite the long distances between the collection sites (from 40.12 to 342.15 Km) and the urban areas and large monocultures around them (which could work as barriers). The capacity to fly long distances (Janzen 1971; Kroodsma 1975; Ackerman et al. 1982), which is one of the best known characteristics of Euglossini bees, could explain the absence of population structure and the similarity of these bee populations. This characteristic can contribute to the maintenance of gene flow, although studies using marking and recapture techniques (Pokorny et al. 2015) or radio telemetry (Wikelski et al. 2010) indicate that the flight range of different species may vary considerably, and we do not have yet these data for the two species used here.
In both species, the grouping can be explained based on phytophysiognomy variation. For Eg. annectans, the populations from EEC and MV that come from the same phytophysiognomy (seasonal semi-deciduous forest) are together in a group and PFBJ, EEJ, and EERP, which came from seasonal semi-deciduous forest with different contact level of Savana biome, form another group. APASJ, which is the only one that belongs to a different phytophysiognomy (montane dense ombrophilous forest), was alone. The dendrograms of morphological similarity, built based on Mahalanobis distance, corroborate the data shown in the scatterplot. For the dendrogram built by Procrustes distance, the grouping pattern was almost the same as in the dendrogram built with Mahalanobis distances. The only exception was EERP, which instead of being alone in one branch, was in the same group with EEC. Discriminant analysis showed a high rate of correct classification; but when we performed the cross-validation test, the rate of identification was somewhat lower, probably based on the high similarity of these samples, as we could see in previous analyses (Mahalanobis and Procrustes distance values).
As for Eg. truncata, the scatterplot built by CVA analysis showed a tendency of sample separation (based on axis 2), where the bees from APASJ were alone and all the other bees were partially superimposed. This aggrupation can be explained based on the same feature used to explain the Eg. annectans data. APASJ is the only site that belongs to montane dense ombrophilous forest, which grouped farther away than all the others, which share seasonal semi-deciduous forest phytophysiognomy. The dendrogram showed an aggrupation that had the same pattern observed in Eg. annectans, except for MV, which grouped with EEJ in the dendrogram built by Mahalanobis distance, and with PFBJ in the dendrogram built based on Procrustes distance. In both cases, the sampling locations share the same phytophysiognomy (seasonal semi-deciduous forest). Discriminant analysis and the cross-validation test showed a high rate of correct classifications in sample identification, similar to the large differences observed in the Mahalanobis and Procrustes distances between groups, compared to the Eg. annectans results.
When we analyzed the dendrograms of morphological similarity, built based on the Mahalanobis and Procrustes distances for all samples of the two species, we also found the same aggrupation pattern, and it followed the phytophysiognomy variation observed in the individual analysis. This aggrupation demonstrated an interesting pattern of environmental traceability, as also reported by Mendes et al. (2007) Francisco et al. (2008), Bischoff et al. (2009), and Francoy et al. (2011) for another bee species, who also used geometric morphometrics analysis of wings. A hypothesis to explain the grouping based on phytophysiognomy variation pattern is phenotypic plasticity, which is the capacity of a single genotype to express a range of phenotypes in response to environmental variation (Fordyce 2006). Phenotypic plasticity is common in insects and it is often highly adaptive (Whitman and Agrawal 2009).
Based on our data, forest fragmentation may not pose a problem for Eg. annectans and Eg. truncata based on an absence of population structure and similarity among the samples from the different sites. The tendency of samples to group by phytophysiognomy could be explained by phenotypic plasticity; according to Via et al. (1995), when environments within the range of a species differ, it may be unlikely that any single phenotype will confer high fitness in all situations. In this case, a change in a phenotype influenced by environment can result in increased environmental tolerance. Phenotypic plasticity could explain adaptation to heterogeneous environments.
References
Ackerman J.D. (1983) Specificity and mutual dependency of the orchid-euglossine bee interaction. Biol. J. Linn. Soc. 20, 301–314.
Ackerman J.D., Mesler M.R., Lu K.L., Montalvo A.M. (1982) Food-foraging behavior of male Euglossini (Hymenoptera: Apidae): vagabonds or trapliners? Biotropica 14, 241–248.
Armbruster W.S., Webster G. (1979) Pollination of two species of Dalechampia (Euphorbiaceae) in Mexico by euglossine bees. Biotropica 11, 278–283.
Bischoff L., Schroeder S., Misof B. (2009) Differentiation and range expansion of North American squash bees Peponapis pruinosa (Apidae: Apiformes) populations assessed by geometric wing morphometry. Ann. Entomol. Soc. Am. 102, 60–69.
Bonatti V., Simões Z.L.P., Franco F.F., Francoy T.M. (2014) Evidence of at least two evolutionary lineages in Melipona subnitida (Apidae, Meliponini) suggested by mtDNA variability and geometric morphometrics of forewings. Naturwissenschaften 101, 17–24
Brook, B.W., Tonkyn, D.W., O’Grady, JJ , Frankham ,R. (2002) Contribution of inbreeding to extinction risk in threatened species. Conserv Ecol 6, http://www.consecol.org/vol6/iss1/art16 (accessed on 17 May 2016)
Brosi B.J. (2009). The effects of forest fragmentation on euglossine bee communities (Hymenoptera: Apidae: Euglossini). Biol. Conserv. 142, 414–423.
Buchmann S.L. (1981) Preliminary anthecological observations on Xiphidium caeruleum Aubl. (Monocotyledonae: Haemodoraceae) in Panama. J. Kansas Entomol. Soc. 53, 685–699.
Cane, J.H. (2001) Habitat fragmentation and native bees: a premature verdict? Conserv Ecol 5 [online] URL: http://www.consecol.org/vol5/iss1/art3/. Accessed 17 May 2016
Cappellari S.C., Harter-Marques B., Aumeier P., Engels W. (2009) Mecardonia tenella (Plantaginaceae) attracts oil-, perfume-, and pollen-gathering bees in Southern Brazil. Biotropica 41, 721–729.
Carnaval A.C., Hickerson M.J., Haddad C.F., Rodrigues M.T., Moritz C. (2009) Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science 323, 785–789.
Cerântola N.D.C.M., Oi C.A., Cervini M., Del Lama M.A. (2011) Genetic differentiation of urban populations of Euglossa cordata from the state of São Paulo, Brazil. Apidologie 42, 214–222
De Souza D.A., Wang Y., Kaftanoglu O., De Jong D., Amdam G.V., Gonçalves L.S., Francoy T.F. (2015) Morphometric identification of queens, workers and intermediates in in vitro reared honey bees (Apis mellifera). PLoS ONE 10, 1–14. Doi:10.1371/journal.pone.0123663
Debat V., Bégin M., Legout H., David J.R. (2003) Allometric and nonallometric components of Drosophila wing shape respond differently to developmental temperature. Evolution 57, 2773–2784.
Dressler R.L. (1982) Biology of the orchid bees (Euglossini). Annu. Rev. Ecol. Syst. 13, 373–394
Eltz T., Schmid M., Roubik D.W. (2003) Fragrances, male display and mating behavior of Euglossa hemichlora—a flight cage experiment. Physiol. Entomol. 28, 251–260.
Eltz T., Sager A., Lunau K. (2005) Juggling with volatiles: exposure of perfumes by displaying male orchid bees. J. Comp. Physiol. A. 191, 575–581.
Fordyce J.A. (2006). The evolutionary consequences of ecological interactions mediated through phenotypic plasticity. J. Exp. Biol. 209, 2377–2383.
Francisco F.O., Nunes-Silva P., Francoy T.M.,Wittmann D., Imperatriz-Fonseca V.L., Arias, M.C., Morgan, E. D. (2008) Morphometrical, biochemical and molecular tools for assessing biodiversity. An example in Plebeia remota (Holmberg, 1903) (Apidae, Meliponini). Insect. Soc. 55, 231–237.
Francoy T.M., Silva R.A.O., Nunes-Silva P., Menezes C., Imperatriz-Fonseca V.L. (2009) Gender identification of five genera of stingless bees (Apidae, Meliponini) based on wing morphology. Genet. Mol. Res. 8, 207–214.
Francoy T.M., Grassi M.L., Imperatriz-Fonseca V.L., de Jesus May-Itza W., & Quezada-Euán J.J.G. (2011). Geometric morphometrics of the wing as a tool for assigning genetic lineages and geographic origin to Melipona beecheii (Hymenoptera: Meliponini). Apidologie 42, 499–507.
Francoy, T.M.,Franco, F.F., Roubik, D.W. (2012). Integrated landmark and outline-based morphometric methods efficiently distinguish species of Euglossa (Hymenoptera, Apidae, Euglossini). Apidologie 43, 609–617.
Frankham R. (2003) Genetics and conservation biology. C. R. Biol. 326, 22–29.
Hurtado-Burillo M., Laura J., May-Itzá W.J., Quezada-Euán J.J., Ruiz C., De la Rúa P. (2016) A geometric morphometric and microsatellite analyses of Scaptotrigona mexicana and S. pectoralis (Apidae: Meliponini) sheds light on the biodiversity of Mesoamerican stingless bees. J. Insect Conserv. 20, 753–763
Janzen D.H. (1971) Euglossine bees as long-distance pollinators of tropical plants. Science 171, 203–205.
Klingenberg C.P. (2011) MorphoJ: an integrated software package for geometric morphometrics. Mol. Ecol. Resour. 11, 353–357.
Knudsen J.T., Mod S.A. (1996) Floral scents and pollination in neotropical Lecythidaceae. Biotropica 28, 42–60.
Kroodsma D.E. (1975) Flight distances of male euglossine bees in orchid pollination. Biotropica 7, 71–72.
Melo G.A. (1995) Fragrance gathering by Euglossa males in flowers of Ternstroemia dentata (Theaceae) (Hymenoptera: Apidae: Euglossinae). Entomol. Gen. 19, 281–283.
Mendes M.F.M., Francoy T.M., Nunes-Silva P., Menezes C., Imperatriz-Fonseca V.L. (2007) Intra-populational variability of Nannotrigona testaceicornis Lepeletier 1836 (Hymenoptera, Meliponini) using relative warp analysis. Biosci. J. 23, 147–152.
Michener C.D. (2000) The bees of the world. The John Hopkins Univ Press, Baltimore, p 913.
Milet-Pinheiro P., Schlindwein C. (2005) Do euglossine males (Apidae, Euglossini) leave tropical rainforest to collect fragrances in sugarcane monocultures? Rev. Bras. Zool. 22, 853–858.
Miller, M.P. (1997) Tools for population genetic analyses (TFPGA): a windows program for the analysis of allozyme and molecular population genetic data. Computer software distributed by author, 4, 157
Myers N., Mittermeier R.A., Mittermeier C.G., Fonseca G.A.B., Kent J. (2000) Biodiversity hotspots for conservation priorities. Nature 403, 853–858.
Nogueira P.C.D.L., Marsaioli A.J., Amaral M.D.C.E., Bittrich V. (1998) The fragrant floral oils of Tovomita species. Phytochemistry 49, 1009–1012.
O’Grady J.J., Reed D.H., Brook B.W., Frankham R. (2008) Extinction risk scales better to generations than years. Anim. Conserv. 11, 442–451.
Oleksa A., Tofilski A. (2014) Wing geometric morphometrics and microsatellite analysis provide similar discrimination of honey bee subspecies. Apidologie 46, 49–60.
Oliveira M.L.D. (2006) Três novas espécies de abelhas da Amazônia pertencentes ao gênero Eulaema (Hymenoptera: Apidae: Euglossini). Acta Amaz 36, 121–128.
Oliveira M.L.D., Nemésio, A. (2003) Exaerete lepeletieri (Hymenoptera: Apidae: Apini: Euglossina): a new cleptoparasitic bee from Amazonia. Lundiana 4, 117–120.
Pokorny, T., Loose, D., Dyker, G., Quezada-Euán, JJG., Eltz, T. (2015) Dispersal ability of male orchid bees and direct evidence for long-range flights. Apidologie 46: 224–237.
Powell A., Powell N.V. (1987) Population dynamics of male euglossine bees in amazonian forest fragments. Biotropica 19, 176–179.
Ramírez S., Dressler R.L., Ospina M.( 2002) Abejas euglosinas (Hymenoptera: Apidae) de la Región Neotropical: Listado de especies con notas sobre su biología. Biota Colombiana 3, 7–118.
Rebêlo J.M.M., Garófalo C.A. (1997) Comunidades de machos de Euglossini (Hymenoptera: Apidae) em Matas Semidecíduas do Nordeste do Estado de São Paulo. Ann. Soc. Entomol. Brasil 26, 243–255.
Rocha-Filho L.C., Cerântola N.C.M., Garófalo C.A., Imperatriz-Fonseca V.L., Del Lama M.A. (2013) Genetic differentiation of the Euglossini (Hymenoptera, Apidae) populations on a mainland coastal plain and an island in southeastern Brasil. Genetica 141, 65–74.
Rohlf F.J. (2008) tpsDig version 2.12. Department of Ecology and Evolution State University of New York Stony Brook
Roubik D., Hanson P. (2004) Orchid bees of tropical America: biology and field guide. Instituto Nacional de Biodiversidad (INBio), Heredia.
Silva F.L., Sella M.L. G., Francoy T.M., Costa A.H.R. (2015). Evaluating classification and feature selection techniques for honeybee subspecies identification using wing images. Comput. Electron. Agric. 114, 68–77.
Siqueira-Filho J.A., Machado I.C. (2008) Flowering phenology and pollination ecology of Cryptanthus dianae Leme: a case of flower fragrance-collecting by Euglossinae bees in Bromeliaceae. Selbyana 29, 226–232.
Soares A.A., Campos L.A.O., Vieira M.F., Melo G.A.R. (1989) Relações entre Euglossa (Euglossella) mandibularis Friese, 1899 (Hymenoptera, Apidae, Euglossini) e Cyphomandra calycina (Solanaceae). Cienc. Cult. 41, 903–905.
Souza R.O., Del Lama M.A., Cervini M., Mortari N., Eltz T., Zimmermann Y., Bach C., Brosi B.J., Suni S., Quezada-Euán J.J.G., Paxton R.J. (2010) Conservation genetics of neotropical pollinators revisited: microsatellite analysis suggests that diploid males are rare in orchid bees. Evolution 64, 3318–3326.
Spielman D., Brook B.W., Frankham R. (2004) Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. 101, 15261–15264.
Suni S.S., Brosi B.J.(2012) Population genetics of orchid bees in a fragmented tropical landscape. Conserv. Genet. 13 323–332.
Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood evolutionary distance and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Teichert H., Dötterl S., Zimma B., Ayasse M., Gottsberger G. (2009) Perfume-collecting male euglossine bees as pollinators of a basal angiosperm: the case of Unonopsis stipitata (Annonaceae). Plant Biol. 11, 29–37.
Tonhasca A., Albuquerque G.S., Blackmer J.L. (2003) Dispersal of euglossine bees between fragments of the Brazilian Atlantic Forest. J. Trop. Ecol. 19, 99–102.
Via S., Gomulkiewicz R., De Jong G., Scheiner S.M., Schlichting C.D., Van Tienderen P.H. (1995) Adaptive phenotypic plasticity: consensus and controversy. Trends Ecol. Evol. 10, 212–217.
Victor, M.A.M. 1975. A devastação florestal. Sociedade Brasileira de Silvicultura. São Paulo, 48p.
Whitman, D.W., Agrawal, A.A. (2009) What is phenotypic plasticity and why is it important? In: Phenotypic plasticity of insects, DW Whitman and TN Ananthakrishnan (Eds.), pp. 1–63
Wikelski M., Moxley J., Eaton-Mordas A., López-Uribe M.M., Holland R., Moskowitz D., Roubik, D.W., Kays, R. (2010) Large-range movements of neotropical orchid bees observed via radio telemetry. PLoS ONE 5: e10738.
Williams N.H., Dressler R.L. (1976) Euglossine pollination of Spathiphyllum (Araceae). Selbyana 1, 349–356.
Zayed A. (2009) Bee genetics and conservation. Apidologie 40, 237–262.
Zimmermann Y., Schorkopf D.L.P., Moritz R.F.A., Pemberton R.W., Quezada-Euan J.J.G., Eltz T. (2011) Population genetic structure of orchid bees (Euglossini) in anthropogenically altered landscapes. Conserv. Genet. 12, 1183–1194.
Acknowledgements
The authors are grateful to FAPESP (Proc. 2011/07857-9 to TMF) and NAP BioComp (University of São Paulo) for financial support. Dr. David de Jong helped us to improve the English of the manuscript.
Author information
Authors and Affiliations
Contributions
MLGS, CAG, and TMF conceived the research and designed experiments; MLGS performed the analysis; MLGS, CAG, and TMF wrote the paper and participated in the revisions of it.
Corresponding author
Additional information
Handling Editor: Marina Meixner
Similarités morphologiques de populations largement séparées de deux espèces d’Euglossini (Hymenoptera; Apidae), basées sur la morphométrie géométrique des ailes
Euglossini / abeilles à orchidées / fragmentation forestière / morphométrie
Morphologische Ähnlichkeit von weit auseinanderliegenden Populationen von zwei Euglossini-Arten (Hymenoptera; Apidae) auf der Basis von geometrischer Morphometrie der Flügel
Euglossini / Prachtbienen / geometrische Morphometrie / Waldfragmentierung
Electronic supplementary material
Supplementary Figure S1
Scatterplot of Euglossa annectans individual scores from the canonical variant analysis of landmarks. See legend to Figure 1 for abbreviations. (GIF 99 kb)
Supplementary Figure S2
Scatterplot of Euglossa truncata individual scores from the canonical variant analysis of landmarks. See legend to Figure 1 for abbreviations. (PNG 73 kb)
Annex
Annex
Supplementary Figure S1
Scatterplot of Euglossa annectans individual scores from the canonical variant analysis of landmarks. See legend to Figure 1 for abbreviations.
Supplementary Figure S2
Scatterplot of Euglossa truncata individual scores from the canonical variant analysis of landmarks. See legend to Figure 1 for abbreviations.
Rights and permissions
About this article
Cite this article
Grassi-Sella, M.L., Garófalo, C.A. & Francoy, T.M. Morphological similarity of widely separated populations of two Euglossini (Hymenoptera; Apidae) species based on geometric morphometrics of wings. Apidologie 49, 151–161 (2018). https://doi.org/10.1007/s13592-017-0536-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-017-0536-0