Abstract
Black queen cell virus (BQCV) is one of the most common honeybee pathogens causing queen brood deaths. The 63 apiaries were sampled between 2007 and 2013 from four different ecogeographic regions in Turkey to estimate BQCV molecular structural characteristics. The BQCV positivity was 47.6%. The 25 local Black queen cell viruses (TrBQCVs) were molecularly characterized and investigated for their genetic relationship with previous records. The identity of the helicase gene among the TrBQCVs was 92–98%, whereas the similarity ranged from 37 to 85% in comparison with the intercontinental records. The identity of the partial capsid gene among the TrBQCVs was 91–100%, and the similarity rate varied from 86 to 97, 88–96, 90–97 and 89–99% in comparison with the Asian, African, American and European counterparts, respectively. The four nonsynonymous substitutions on the partial capsid protein suggest a predicted genotype that is specific among TrBQCVs.
Similar content being viewed by others
1 Introduction
Honeybees are economically predominant pollinator insects and serve to enhance the biodiversity of the ecosystem. Beekeeping and bee products are critically important in the socioeconomic world of global food suppliers. Honeybee colony health has gained importance because they are currently threatened by sublethal exposure to pesticides, parasites, pests, poor nutrition and pathogens, including viruses, bacteria and fungi. Honeybee viruses also cause covert and overt infections and have the potential to cause considerable economic problems for apiculture by the adverse influence on pupae, larvae and adults (Hails et al. 2008; Dainat et al. 2012).
Black queen cell virus (BQCV) belongs to the Cripavirus genus within the Dicistroviridae family (Mayo 2002). The viral genome consists of a single-stranded RNA molecule that is 8550 nt long. Two open reading frames separate the RNA molecule (ORFs). ORF-1, which is located in the 5’proximal region, encodes a replicase polyprotein that includes helicase, RNA-dependent RNA polymerase (RdRp) and 3C-like cysteine protease, whereas the 3’proximal ORF-2 encodes capsid polyprotein (Leat et al. 2000). BQCV is one of the most widespread and commonly isolated honeybee pathogens (Berenyi et al. 2006) since it was first reported in 1955. It was first isolated from queen, prepupae, pupae are characterized by darkening. In addition, what appear to be healthy and persistent adult bees can be subclinically infected (Bailey and Woods 1977).
Beekeeping is an important sector for Turkey. The sudden colony losses that have been reported since 2006 are similar to many countries in the world and have been an ongoing and primary concern for Turkey. Previously, reported BQCV prevalence in Turkey was 21.42% (Gumusova et al. 2010), but there is no information regarding the molecular structures of BQCV genotypes in Turkey. The molecular characterization and phylogenetic analysis of TrBQCV genotypes was performed in this study.
2 Materials and methods
2.1 Samples
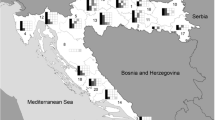
Honeybee samples (A. mellifera L.) were collected in four different ecogeographic regions from the Nortwest, South, Middle and Southeast of Turkey (Figure 1). Sampling was conducted at locations of significant apiculture and at national queen bee breeders. All the samples were collected from beekeepers who complained sudden colony losses between 2007 and 2013. A total of 126 colonies were sampled from 63 apiaries, and two colonies from each apiary were sampled by collecting ten worker bees each. All the sampled queens and worker bees were seemingly healthy, but the pupae and larvae showed BQCV signs. A total of 35 queen bee nucs were sampled. The number of the bees tested in this study were based on beekeeper submission and after delivered to laboratory, they were stored at − 80 °C until ready for use.
2.2 RNA extraction and RT-PCR
The honeybee samples were homogenized individually in 0.5 mL sterile phosphate-buffered cold solution (Sigma, USA) and cold centrifuged at 14000×g for 5 min. The total RNA was isolated from the supernatant using two methods performed according to the sampling date. The acid guanidinium thiocyanate-phenol-chloroform extraction method, which was previously described by Chomczynski and Sacchi (2006), was performed for honeybee samples collected from 2007 and 2009. TRIzol solution (Invitrogen, CA) was used according to the manufacturer’s instructions for samples collected between 2010 and 2013. The total RNA was eluted in a final volume of 30 μL for both methods, and ribonuclease inhibitor was added (RNaseOUT, Invitrogen, CA) at 10% (v/v) to the tube. All the purified RNA samples were stored at − 80 °C for further analysis. Reverse transcription was conducted using a commercial cDNA synthesis kit (First Strand cDNA Synthesis Kit, Fermentas, Lithuania) with random hexamer primers following the manufacturer’s recommendations. The BQCV capsid protein coding the gene region and the nonstructural polyprotein-helicase enzyme-coding gene region were targeted in PCR. For this purpose, previously reported primer pairs (Benjeddou et al. 2001; Berenyi et al. 2006) were used and are listed in Figure 2.
Primer pairs were used for PCR amplification and sequencing of BQCV genotypes in this study. Asterisk: the amplified regions in PCR in this study. Number sign: nucleotide position refers to the published BQCV genome sequences including accession number AF125252 for nonstructural polyprotein gene and accession number AF183905 for capsid gene region.
The PCR mixtures for each sample included 5 μL of cDNA template (~ 0.1 μg), 10× PCR buffer, 3 mM MgCl2, 0.2 mM dNTP mix, 0.3 μM of each primer and 2 U hot start Taq polymerase (Maxima Hot Start Taq DNA Polymerase, Fermentas). The thermal cycling profiles were as follows: an initial denaturation at 95 °C for 5 min, then 40 cycles of denaturation at 94 °C for 1 min, an annealing temperature at 48 °C (BQCV-3f-4r primer pair) and at 60 °C (BQCV-J1-J2 primer pair) for 1 min, and extension at 70 °C for 1 min. The heat was increased to 72 °C for 7 min at the final extension. Water was used as the negative control and positive control (detected previously) in each run of RT-PCR and the PCR protocols. All the amplified products were electrophoresed in 1.5% agarose gel containing 0.5 μg/mL EtBr and DNA bands visualized with a UV transilluminator (UVP Inc., USA).
2.3 Nucleotide sequencing and phylogenetic analysis
Specific PCR products were excised from the agarose gel and purified using a commercial purification kit (GelElute Gel Extraction Kit, Sigma, USA) according to the manufacturer’s instructions. Sequence analysis based on the Sanger method was performed using an Applied Biosystems DNA analyser. Nucleotide sequences were compared and identified using a Basic Local Alignment Search Tool (BLAST) web program (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch). The alignment of each fragment was conducted using the BioEdit program (v.7.2.5) (Hall 1999). A phylogenetic analysis was generated by the neighbour-joining and maximum-likelihood methods with bootstrapping of 1000 replicates using MEGA 6.1 software (Tamura et al. 2013). The nucleotide sequences were translated into amino acid sequences using the Expasy tool (http://web.expasy.org/translate/). The distance matrix, p distance analysis and similarity level for each fragment sequence were investigated along with previously reported BQCV genotypes (Leat et al. 2000; Reddy et al. 2013; Mookhploy et al. 2015). An already completed South African BQCV full genome (accession number of AF183905) was used as the reference genotype (Leat et al. 2000). The deduced amino acid of the partial capsid gene region was evaluated regarding positively and negatively selected sites using the HyPhy package (Datamonkey, http://www.datamonkey.org). The cut-off criterion of the p value was set at 0.1.
2.4 Accession numbers of nucleotide sequences
A total of the 25 BQCV sequences were obtained and identified and were submitted to the GenBank database with the accession numbers FJ588532-FJ588535 for the nonstructural polyprotein coding gene region and the accession numbers KX273080-KX273100 for the capsid gene region.
3 Results
3.1 RT-PCR
From 2007 to 2013, all bee samples were examined for viral genomes by the RT-PCR assay. The BQCV positivity was 47.6% and ranged between 22.2 and 100% among the apiaries (Table I). A total of four seemingly healthy queen bees, 18 worker bees, two larvae and a pupae sample that were found to be BQCV positive were used for sequencing.
3.2 Genetic variability of TrBQCVs
The 25 nucleotide sequences, which included four partial nonstructural polyprotein-helicase enzyme-coding gene regions (ORF-1) and 21 partial capsid protein coding gene regions (ORF-2), were identified. The distributions of the p distance value among TrBQCVs varied from 0.018–0.054 (mean 0.036) and 0.003–0.054 (mean 0.035) for the helicase gene and capsid gene regions, respectively. TrBQCVs showed 92–98% identity to each other based on the helicase coding gene region, while sequence similarity with previous sequences was 37–85%. Among the queen bee-originated helicase gene sequences (TR-HA13 and TR-15A), a high identity rate was found with a similarity of 98%. In the comparative sequence analysis, TrBQCVs were closer to European sequences (81–85%) than the South African sequences (79–81%). When comparing TrBQCVs to Asian sequences, the similarity rate variability was 37–81% with Chinese sequences and 37–39% with South Korean viruses. The partial capsid gene sequences were found to be 91–100% identical among the TrBQCVs. They shared similarity between Asian, African, American and European origin genotypes with rate of 86–97, 88–96, 90–97 and 89–99%, respectively. The UK isolate (AF125252) among the European sequences was closer (94–99%) to TrBQCVs wherease the Chinese genotype (AB746348; 86–93%) from the Asian sequences and the Uruguay genotype (DQ364629; 88–93%) from the South America genotypes showed less similarity with the TrBQCVs.
At the amino acid level, the identity rate of the helicase C domain coding gene was 98–99% among the TrBQCVs, whereas this rate varied from 90 to 93% with European and 78–79% with Asian genotypes. Two of the more variable regions were highly conserved among the TrBQCVs and were found in multiple sequence alignments (Figure 3). They also shared some common points with the UK isolate (AF125252).The identity rate of the deduced amino acid of the TrBQCVs partial capsid protein was 97–100%, and 92–100% with other selected viruses. The TrBQCVs had a lower similarity rate compared with the Chinese (AB746348; 92–94%), Korean (JX149518; 93–94%) and Uruguay (DQ364629; 93–95%) viruses. The TrBQCVs showed variability in four regions according to multiple alignment sequences and compared to the reference (data not shown).
Multiple sequence alignment of partial gene coding Helicase enzyme (domain C) region among TrBQCVs in this study. The sequences compared with other sequences from the different continent which South Africa (as reference), UK, Poland, Hungary, China. The common variable regions among sequences were indicated by the line.
The positive and negative pressure selection sites on the partial capsid gene region among the TrBQCVs were estimated by codon-based methods. The five selected sites were found in the SLAC (p = 0.058), FEL (p = 0.022–0.078) and IFEL (p = 0.021–0.084) analysis, whereas many sites were under negative selection. Of them, only one positive selected substitution was estimated as a specified genotype (Y689F) when compared to the phylogenetic tree. Furthermore, some amino acid differences were defined on the partial capsid protein among the TrBQCVs (Table II). In particular, the four amino acid substitutions at Y689F, T780S, V781I and T839A were predicted to be genotype specified sites and would be effective in the phylodynamic of the TrBQCVs (Figure 5). Another codon found as one of the negative pressure selected locations (719) in this study shared a common site among Asian BQCV genotypes and were different from the others (altered as D719E).
3.3 Phylogenetic analysis of the TrBQCVs
The phylogenetic tree was constructed with the genes as mentioned above. For the helicase gene, the tree reflected two main clusters in which there was one Asian genotype and other viruses (Figure 4). The TrBQCVs were highly conserved within and slightly separated from the other strains. They composed a cluster within one of the main groups in the tree and were entirely different from the conserved subgroup.
Phylogenetic analysis of helicase gene regions of TrBQCV nucleotide sequences isolated from various parts of Turkey. The phylogenetic tree included the sequences selected from the different countries aligned with Turkish sequences was constructed by Mega 6.1 program (Tamura et al. 2013) by using the maximum-likelihood method with Kimura’s two-parameter model and 1000 replicates bootstrap value (only node value above 50% are shown). The red dot indicates TrBQCVs.
The phylogenetic tree based on the partial capsid gene among the TrBQCVs was clustered by two main groups as group I and group II. One of them shared common amino acid differences at some substitutions (Figure 5). The nine members of group II (excluding TrBQCV-14) had common substitution differences at V781I. TrBQCV-1, TrBQCV-13, TrBQCV-16 and TrBQCV-20 shared both T780S and V781I differences; they also were clustered in the same branch in the phylogenetic tree. In addition, the results showed that the TrBQCVs were placed in the phylogenetic tree independently from their isolation date. In the phylogenetic comparison (Figure 7), the TrBQCVs were separated from the other sequences, excluding the UK and Germany sequences. The UK sequences were placed within group I, whereas the Germany sequence was within the group II cluster. The Asian sequences (China, Japan, Korea, South Korea, Thailand and Taiwan) formed a common cluster including a US isolate distinct from TrBQCVs. The predicted three-dimensional structures of the viral partial capsid gene for the TrBQCVs were generated using the SWISS-MODEL server (Figure 6). Among all the target sequences, two preliminary models were inferred as best homology selections. Three points, excluding overlap models, were suggested as differences and comparable sites for protein conformation.
Phylogenetic analysis of partial capsid gene regions of TrBQCV nucleotide sequences isolated from different parts of Turkey. The phylogenetic tree was constructed by Mega 6.1 program (Tamura et al. 2013) by using the maximum-likelihood method with Kimura’s two-parameter model and 1000 replicates bootstrap value (only node value above 50% are shown). The amino acid changes among isolates were shown. The branch length of nucleotide substitutions per site was used 0.005 bar.
4 Discussion
Turkey is one of the world’s largest beekeeping countries located between Europe, Caucasia and Asia, and this permits her to become an intercontinental biogenetic switch/exchange in biogeography. There are 64,500 registered beekeepers and 7,528,000 hives in Turkey (TurkVet 2016). Since 2006, beekeepers have been complaining about colony level and unusual disease scenarios resulting in progressive declines and severe losses. The colony losses have been mentioned by stationary or migratory beekeepers as well as in queen rearing apiaries. The individual reasons of colony depopulations in Turkey are certainly unknown, but infected honeybee colonies with various pathogenic strains may become chronic carriers and source of economically important latent and persistent infections for colony health (Reddy et al. 2013). Few studies have focused on viral honeybee pathogens in Turkey (Gumusova et al. 2010; Muz and Muz 2009; Tozkar et al. 2015).The BQCV positivity was reported from the apiaries sampled in 2009 (Gumusova et al. 2010) and between 2010 and 2011 years (Tozkar et al. 2015), but there is still no detail about the molecular structure of TrBQCVs. The samples used in this study were collected from 2007 to 2013 and after a beekeeper’s extraordinary reports regarding serious honeybee losses. The average positivity of BQCV was 47.6%, and the positivity rate varied from 22.2 to 100% in the current study.
Researchers have estimated some hypothetical evolutionary scenarios for positive-sense RNA viruses. It has hypothesised that all positive-strand RNA viruses could have emerged from a common ancestor virus that contained genes for RNA-dependent RNA polymerase, a protease that also acted as the capsid protein, and an RNA helicase. However, these gene regions were focused on by scientists for the purpose of the taxonomy of positive-sense RNA viruses, including honeybee viruses (Koonin et al. 1993; Tapaszti et al. 2009; Singh et al. 2010; Mookhploy et al. 2015; Ravoet et al. 2015). The partial helicase gene region coding the helicase C domain and the partial capsid gene region were examined with regard to genetic divergence and molecular differences among the Turkish field viruses. The helicase enzyme-coding gene region was more variable than the capsid protein coding gene region at the nucleotide level, whereas it was partially conserved at the amino acid level. The differences and possible separations among the field viruses were also successfully screened in the phylogenetic trees. It is possible that the genetic diversity and mutations within the conserved genes of the field viruses could emerge.
Information regarding the nucleotide sequence and genetic classification of BQCV genotypes is globally very limited (Leat et al. 2000; Benjeddou et al. 2001; Tapaszti et al. 2009; Singh et al. 2010; Mookhploy et al. 2015; Ravoet et al. 2015). In the current study, the two viral genes were examined and the current results show that TrBQCVs highly conserved with each other and displayed a short genetic distance for the helicase gene (p value = 0.036) and the capsid gene (p value = 0.035). They exhibited a more variable rate with other selected viruses from different continents according to multiple sequence alingments. They are placed in a different cluster in the phylogenetic tree-based on helicase gene (Figure 4). Regarding the capsid gene, the TrBQCVs mainly clustered as divergent from others while they were sub-divided into two groups among themselves in the phylodynamic (Figures 5 and 7). The phylogenetic status of all the sequences seemed generally corresponding to their geographical origins. The similarity rate of TrBQCVs with UK, China, Uruguay and South Africa genotypes is 94–99, 86–93, 88–93 and 88–96%, respectively. The homology among the genotypes was compatible with their phylogenetic distributions. The accordance between the results of homology and phylogenetic analysis may highlight the possibility to predict the origins of field genotypes in future.
Phylogenetic analysis of partial capsid gene regions of BQCV nucleotide sequences isolated from different parts of Turkey. The phylogenetic tree included the sequences obtained from the different countries aligned with Turkish sequences was constructed by Mega 6.1 program (Tamura et al.2013) by using the maximum-likelihood method with Kimura’s two-parameter model and 1000 replicates bootstrap value (only node value above 50% are shown). TrBQCVs are indicated by the red dot.
The ORF1 and ORF2 regions of the BQCV genotypes according to the geographic location of the honeybee have also been discussed for genetic researches of field viruses by several studies (Tapaszti et al. 2009; Noh et al. 2013; Reddy et al. 2013). In general, phylogenetic analyses of field viruses were performed based on the ORF2 gene (Tapaszti et al. 2009; Reddy et al. 2013; Mookhploy et al. 2015; Tsevegmid et al. 2016). Nevertheless, TrBQCVs had differences in phylogenetic classification for both the studied gene regions. TrBQCVs were even sub-divided into different clusters, including one group for the helicase gene and two groups for the capsid gene; however, they were closer to some central European viruses among the selected viruses based on the capsid gene. The ORF2 gene region could be more practical to investigate the genetic relationship of field viruses and the viral molecular structure. In addition, in comparison with different countries of origin, the sequences of Deformed wing virus (Berenyi et al. 2006) and Sacbrood virus (Choe et al. 2012) strains were reported to show divergence according to their origins as well as the BQCV sequences (Koonin et al. 1993; Tapaszti et al. 2009). The current results also showed that the geographic origins of global sequences would be decisive for their genetic divergence. The BQCV infected queen bees can transfer the virus from one generation to next generation (Chen et al. 2006). This may explain the global impact of the queen bee health for apiaries.
The capsid protein of the Picornaviridae family covers major antigenic sites of the virus and can affect the viral function and dynamic host immunity (Leat et al. 2000). We thought that it could be evaluated as antigenic sites associated with selective pressure in similar host mammalian viruses (Hirano et al. 2014). Therefore, positive and negative selection sites were analysed for the partial capsid protein of TrBQCVs (Figures 5 and 6). Many negative selection sites and few positive selection sites were found, and one positive site interpreted as Y689F would be especially useful for phylogenotyping of field strains. Moreover, three amino acid substitutions, T780S, V781I and T839A, were evaluated to predict the genotype-specific sites among the TrBQCVs. These changes were not placed on other strains, but most Asian viruses shared the D719E substitution.
Homology models of the virion structure are useful to predict epitope sites and could be used to infer the virion surface and then targeted by host immunity. We have limited information regarding insect host-virus interaction, especially in virus escape strategies from the host immunity and fighting between the host and virus. To design an antiviral strategy, drug or vaccine against viral infection, there may need to be similar approaches (Susevich et al. 2015; Ekins et al. 2016; Kalynych et al. 2016). Therefore, conformational models are of major importance and help predict the target epitope site. The predicted three-dimensional structures of the viral partial capsid gene for TrBQCVs were generated in this study (Figure 6) and two preliminary models were inferred as best homology selections. Three points excluding overlap models were suggested as differences and comparable sites for protein conformation.
To have healthy bees in a colony is significantly important for the sustainability of the colony. Increased mortality in the brood and queen cells threatens the colony health and leads to significant economic damage in an apiary. Honeybee pathogens, particularly viruses, are important causative agents affecting brood health (Hails et al. 2008). BQCV causes brood health problems with risk of colony sustainability (Dainat et al. 2012). The field originated phylogenetic examination of BQCV genotype divergence may help to understand global epidemiology of the disease.
The current results indicate that the geographic origin differences of global BQCV genotypes could be useful for phylogenetic clustering. Finally TrBQCVs were more conserved among each other and separate from most of different BQCV sequences. This study reveals the first data regarding the molecular genetic structures of TrBQCVs and will be useful for further research.
References
Bailey, L., Woods, R.D. (1977) Two more small RNA viruses from honey bees and further observations on sacbrood and acute bee-paralysis viruses. J. Gen. Virol. 25, 175–186
Benjeddou, M., Leat, N., Allsopp, M., Davison, S. (2001) Detection of acute bee paralysis virus and black queen cell virus from honeybees by reverse transcriptase PCR. Appl. Environ. Microb. 67, 2384–2387
Berenyi, O., Bakonyi, T., Derakhshifar, I., Koglberger, H., Nowotny, N. (2006) Occurrence of six honeybee viruses in diseased Austrian apiaries. Appl. Environ. Microbiol. 72 (4), 2414–20
Chen, Y.P., Evans, J.D., Feldlaufer, M. (2006) Horizontal and vertical transmission of viruses in the honeybee, Apis mellifera. J. Invertebr. Pathol. 92, 152–159
Choe, S.E., Nguyen, L.T., Noh, J.H., Koh, H.B., Jean, Y.H., Kweon, C.H., Kang, S.W. (2012) Prevalence and distribution of six bee viruses in Korean Apis cerana populations. J Invertebr. Pathol. 109(3), 330–3
Chomczynski, P., Sacchi, N. (2006) The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nat. Protoc. 1, 581–585
Dainat, B., Evans, J.D., Chen, Y.P., Gauthier, L., Neumann, P. (2012) Predictive markers of honey bee colony collapse. PLoS One7(2), e32151
Ekins, S., Liebler, J., Neves, B.J., Lewis, W.G., Coffee, M., Bienstock, R., Southan, C., Andrade, C. CH. (2016) Illustrating and homology modeling the proteins of the Zika virus. F1000Research. 5, 275 [version 2; referees: 2 approved].
Gumusova, O., Albayrak, H., Kurt, M., Yazici, Z. (2010) Prevalence of three honey bee viruses in Turkey. Vet. Arh. 80,779–785
Hall, T.A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41:95–98.
Hails R.S., Ball B.V., Genersch E., (2008). Infection strategies of insect viruses, in: Aubert M., Ball B.V., Fries I., Moritz R.F.A., Milani N., Bernardinelli I. (Eds.), Virology and the honey bee, European Communities, Luxembourg, pp. 255–275. http://www.eurosfaire.prd.fr/7pc/doc/1262083725_virology_and_the_honey_bee.pdf.
Hirano, E., Kobayashi, M., Tsukagoshi, H., Yoshida, L.M., Kuroda, M., et al. (2014) Molecular evolution of human respiratory syncytial virus attachment glycoprotein (G) gene of new genotype ON1 and ancestor NA1. Infect. Genet. Evol. 28, 183–91
Kalynych, S., Pridal, A., Palkova, L., Levdansky, Y., de Miranda, J.R., Plevka, P. (2016) Virion structure of iflavirus slow bee paralysis virus at 2.6-angstrom resolution. J. Virol.. 90,7444–7455
Koonin, E.V., Dolja, V.V, Morris, T.J. (1993) Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol.. 28 (5), 375–430
Leat, N., Ball, B., Govan, V., Davison, S. (2000) Analysis of the complete genome sequence of black queen cell virus, a picorna-like virus of honey bees. J. Gen. Virol. 81, 2111–2119
Mayo, M.A. (2002) Virus taxonomy—Houston. Arch. Virol. 147 (5),1071–6
Mookhploy, W., Kimura, K., Disayathanoowat, T., Yoshiyama, M., Hondo, K., Chantawannakul, P. (2015) Capsid gene divergence of black queen cell virus isolates in Thailand and Japan honey bee species. J. Econ. Entomol. 108 (3), 1460–4
Muz, D., Muz, M.N. (2009) Survey of the occurrence of deformed wing virus and multiple parasites of queens (Apis mellifera L.) in apiaries with collapsed colonies in Hatay, Turkey. J. Apic. Res., 48 (3), 204–208
Noh, J.H., Reddy, K.E., Choe, S.E., Yoo, M.S., Doan, H.T., et al. (2013) Phylogenetic analysis of black queen cell virus genotypes in South Korea. Virus Genes 46 (2),362–8
Ravoet, J., De Smet, L., Wenseleers, T., de Graaf, DC. (2015) Genome sequence heterogeneity of Lake Sinai Virus found in honey bees and Orf1/RdRP-based polymorphisms in a single host. Vir Res 201, 67–72
Reddy, K.E., Noh, J.H., Choe, S.E., Kweon, C.H., Yoo, M.S., et al. (2013) Analysis of the complete genome sequence and capsid region of black queen cell viruses from infected honeybees (Apis mellifera) in Korea. Virus Genes 47, 126–132
Singh, R., Levitt, A.L., Rajotte, E.G., Holmes, E.C., Ostiguy, N., vanEngelsdorp, D., Lipkin, W.I., Depamphilis, C.W., Toth, A.L., Cox-Foster, D.L. (2010) RNA viruses in hymenopteran pollinators: evidence of inter-taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS ONE 5, e14357.
Susevich, M.L., Marti, G.A., Balsalobre, A., Echeverria, M.G. (2015) Phylogenetics based on partial ORF2 of triatoma virus in triatomines collected over a decade from domiciliary habitats. J. Insect Sci.. 15(1), 17
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013) PMCID: PMC3840312 MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 (12), 2725–2729
Tapaszti, Z., Forgách, P., Kővágó, C., Topolska, G., Nowotny, N., Rusvai, M., Bakonyi, T. (2009) Genetic analysis and phylogenetic comparison of Black queen cell virus genotypes. Vet Mic 139(3–4), 227–234
Tozkar, C.O., Kence, M., Kence, A., Huang, Q., Evans, J.D. (2015) Metatranscriptomic analyses of honey bee colonies. Front. Genet. 6, 100
Tsevegmid, K., Neumann, P., Yañez, O. (2016) The honey bee pathosphere of Mongolia:European viruses in Central Asia. PLoS One. 11(3):e0151164.
TurkVet, (2016). http://www.turkvet.gov.tr/. Accession date: 12/14/2016
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Contributions
DM: participated in design and interpretation experiments and analysis, wrote the paper; MNM DM: participated in design and interpretation, performed experiments and analysis, wrote the paper.
Additional information
Manuscript editor: Klaus Hartfelder
Une étude épidémiologique moléculaire du virus de la cellule royale noire chez les abeilles ( Apis mellifera ) de Turquie: la première caractérisation génétique et analyse phylogénétique de virus isolés à partir de différents ruchers
abeille / BQCV / gène des protéines de la capside / gène hélicase / Turquie
Eine molekularepidemiologische Untersuchung des Black Queen Cell Virus in Honigbienen ( Apis mellifera ) der Türkei: Eine erste genetische Charakterisierung und phylogenetische Analyse von Viren aus Feldproben
Honigbiene / BQCV / Helikase-Gen / Türkei
Rights and permissions
About this article
Cite this article
MUZ, D., MUZ, M.N. A molecular epidemiological study of black queen cell virus in honeybees (Apis mellifera) of Turkey: the first genetic characterization and phylogenetic analysis of field viruses. Apidologie 49, 89–100 (2018). https://doi.org/10.1007/s13592-017-0531-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-017-0531-5