Abstract
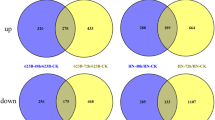
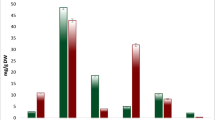
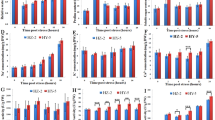
Tartary buckwheat (Fagopyrum tataricum) is widely planted in the world because of its high nutritional content and strong salt tolerance. However, high salinity still causes some declines in the crop production. In addition, it is unclear of molecular mechanism imparting salt tolerance on this crop. In order to obtain more detailed information regarding this mechanism, we made transcriptome comparisons between short-term acclimation to salt stress (100 mM of NaCl, 48 h) and control in two existing and two new salt-tolerant cultivars of Tartary buckwheat in this study. In total, 205.2 million clean reads were produced through an illumina sequencing approach. A total of 10,865 unigenes were annotated against all four public databases: Nr, Swiss-Prot, KOG and KEGG. 42 common unigenes in all the four cultivars expressed differentially after 100 mM of NaCl for 48 h, suggesting that these differentially expressed unigenes (DEGs) may be involved in salinity response. In addition, we identified 57 transcription factor families total in this study, whereas eight of 42 DEGs can encode this protein. We also observed certain unigenes were ROS (reactive oxygen species) related, including superoxide dismutase (SOD), peroxidase (POD), and ascorbate peroxidase (APX), which were chosen for validation by quantitative real-time PCR. Taken together, the transcriptome data obtained from distinct cultivars of F. tataricum with different salt tolerance was the first report, which provides a valuable insight into the mechanisms of salt tolerance and offers promising references for cultivating strong salt-tolerant Tartary buckwheat cultivars.










Similar content being viewed by others
Data availability
Raw reads have been deposited to the NCBI Sequence Read Archive under the accession ID SRP125065: https://www.ncbi.nlm.nih.gov/sra/?term=SRP125065.
Abbreviations
- CDS:
-
Coding sequence
- DEG:
-
Differentially expressed gene
- FDR:
-
False discovery rate
- GO:
-
Gene ontology
- MAPK:
-
Mitogen activated protein kinase
- ROS:
-
Reactive oxygen species
- SOS:
-
Salt overly sensitive
- SR:
-
Salt-related
- SRA:
-
Sequence read archive
- TF:
-
Transcription factor
References
Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13(3):99–102. https://doi.org/10.1016/j.tplants.2007.11.012
Askira Y, Rubin B, Rabinowitch HD (1991) Differential response to the herbicidal activity of delta-aminolevulinic acid in plants with high and low SOD activity. Free Radic Res Commun 13(1):837–843. https://doi.org/10.3109/10715769109145865
Azooz MM, Ismail AM, Elhamd MFA (2009) Growth, lipid peroxidation and antioxidant enzyme activities as a selection criterion for the salt tolerance of maize cultivars grown under salinity stress. Int J Agric Biol 11:572–577. https://doi.org/10.15439/2014F360
Belver A, Olias R, Huertas R, Rodriguez-Rosales MP (2012) Involvement of SlSOS2 in tomato salt tolerance. Bioengineered 3:298–302. https://doi.org/10.4161/bioe.20796
Butcher K, Wick AF, Desutter T, Chatterjee A, Harmon J (2016) Soil salinity: a threat to global food security. Agron J 108:2189–2200. https://doi.org/10.2134/agronj2016.06.0368
Büyük İ, Aydın SS, Duman DC, Aras S (2014) Expression analysis of APX and CAT genes in eggplants subjected to Cu+2 and Zn+2 heavy metals. New Biotechnol 31:S138. https://doi.org/10.1016/j.nbt.2014.05.1956
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. https://doi.org/10.1093/bioinformatics/bti610
Dai X, Sinharoy S, Udvardi M, Zhao PX (2013) PlantTFcat: an online plant transcription factor and transcriptional regulator categorization and analysis tool. BMC Bioinform 14:321. https://doi.org/10.1186/1471-2105-14-321
Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K (2007) Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 143:1739–1751. https://doi.org/10.1016/S0168-8278(08)60689-3
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573. https://doi.org/10.1016/j.tplants.2010.06.005
Esfandiari E, Gohari G (2017) Response of ROS-scavenging systems to salinity stress in two different wheat (Triticum aestivum L.) cultivars. Not Bot Horti Agrobo 45(1):287–291. https://doi.org/10.15835/nbha45110682
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10:366–371. https://doi.org/10.1016/j.pbi.2007.04.020
Fabjan N, Rode J, Košir IJ, Wang Z, Zhang Z, Kreft I (2003) Tartary buckwheat (Fagopyrum tataricum Gaertn.) as a source of dietary rutin and quercitrin. J Agric Food Chem 51:6452–6455. https://doi.org/10.1021/jf034543e
Fang X, Huang K, Nie J, Zhang Y, Yi Z (2019) Genome-wide mining, characterization, and development of microsatellite markers in Tartary buckwheat (Fagopyrum tataricum Garetn.). Euphytica 215:183. https://doi.org/10.1007/s10681-019-2502-6
Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9:436–442. https://doi.org/10.1016/j.pbi.2006.05.014
Ganesan G, Sankararamasubramanian HM, Narayanan JM, Sivaprakash KR, Parida A (2008) Transcript level characterization of a cDNA encoding stress regulated NAC transcription factor in the mangrove plant Avicennia marina. Plant Physiol Biochem 46:928–934. https://doi.org/10.1016/j.plaphy.2008.05.002
Gossett DR, Millhollon EP, Lucas MC, Banks SW, Marney MM (1994) The effects of NaCl on antioxidant enzyme activities in callus tissue of salt-tolerant and salt-sensitive cotton cultivars (Gossypium hirsutum L.). Plant Cell Rep 13:498–503. https://doi.org/10.1007/bf00232944
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Gu J, Huang LX, Gong YJ, Zheng SC, Liu L, Huang LH, Feng QL (2013) De novo characterization of transcriptome and gene expression dynamics inepidermis during the larval-pupal metamorphosis of common cutworm. Insect Biochem Mol Biol 43:794–808. https://doi.org/10.1016/j.ibmb.2013.06.001
Iseli C, Jongeneel CV, Bucher P (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol 99:138–148. https://doi.org/10.1002/9780470999455.ch1
Jaiswal P (2013) Maize metabolic network construction and transcriptome analysis. Plant Genome 6:1–12. https://doi.org/10.3835/plantgenome2012.09.0025
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. https://doi.org/10.1093/nar/28.1.27
Khan SA, Li MZ, Wang SM, Yin HJ (2018) Revisiting the role of plant transcription factors in the battle against abiotic stress. Int J Mol Sci 19(6):1634. https://doi.org/10.3390/ijms19061634
Koonin EV, Fedorova ND, Jackson JD et al (2004) A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Boil 5:R7. https://doi.org/10.1186/gb-2004-5-2-r7
Krulwich TA (1983) Na+/H+ antiporters. Biochim Biophys Acta 726:245–264. https://doi.org/10.1016/0304-4173(83)90011-3
Kuimelis RG, Livak KJ, Mullah B, Andrus A (1997) Structural analogues of TaqMan probes for real-time quantitative PCR. Nucleic Acids Symp Ser 37:255–256
Letey J, Hoffman GJ, Hopmans JW, Grattan SR, Suarez D, Corwin DL, Oster JD, Wu L, Amrhein C (2011) Evaluation of soil salinity leaching requirement guidelines. Agric Water Manag 98:502–506. https://doi.org/10.1016/j.agwat.2010.08.009
Li D, Su Z, Dong J, Wang T (2009a) An expression database for roots of the model legume Medicago truncatula under salt stress. BMC Genomics 10:517. https://doi.org/10.1186/1471-2164-10-517
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009b) SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25(15):1966–1967. https://doi.org/10.1093/bioinformatics/btp336
Little JW, Mount DW (1982) The SOS regulatory system of Escherichia coli. Cell 29:11–22. https://doi.org/10.1016/0092-8674(82)90085-X
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Ma S, Gong Q, Bohnert HJ (2006) Dissecting salt stress pathways. J Exp Bot 57:1097–1107. https://doi.org/10.1093/jxb/erj098
Makinen V, Salmela L, Ylinen J (2012) Normalized N50 assembly metric using gap-restricted co-linear chaining. BMC Bioinform 13(1):255. https://doi.org/10.1186/1471-2105-13-255
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793. https://doi.org/10.2307/1592215
Metternicht GI, Zinck JA (2003) Remote sensing of soil salinity: potentials and constraints. Remote Sens Environ 85:1–20. https://doi.org/10.1016/S0034-4257(02)00188-8
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182-185. https://doi.org/10.1093/nar/gkm321
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. https://doi.org/10.1038/nmeth.1226
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59(1):651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
Nagalakshmi U, Waern K, Snyder M (2010) RNA-seq: a method for comprehensive transcriptome analysis. Curr Protoc Mol Biol 4:11–13. https://doi.org/10.1002/0471142727.mb0411s89
Nakashima K, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149:88–95. https://doi.org/10.1104/pp.108.129791
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87. https://doi.org/10.1016/j.tplants.2004.12.010
Pang CH, Wang BS (2008) Oxidative stress and salt tolerance in plants. Prog Bot 69:231–245. https://doi.org/10.1007/978-3-540-72954-9_9
Peng FY, Weselake RJ (2013) Genome-wide identification and analysis of the B3 superfamily of transcription factors in Brassicaceae and major crop plants. Theor Appl Genet 126:1305–1319. https://doi.org/10.1007/s00122-013-2054-4
Pitman MG, Lauchli A (2002) Global impact of salinity and agricultural ecosystems. Environ Plants Mol 1:3–20. https://doi.org/10.1007/0-306-48155-3_1
Qiao WH, Zhao XY, Li W, Luo Y, Zhang XS (2007) Overexpression of AeNHX1, a root-specific vacuolar Na+/H+ antiporter from Agropyron elongatum, confers salt tolerance to Arabidopsis and Festuca plants. Plant Cell Rep 26:1663–1672. https://doi.org/10.1007/s00299-007-0354-3
Rahman H, Ramanathan V, Nallathambi J, Duraialagaraja S, Muthurajan R (2016) Over-expression of a NAC 67 transcription factor from finger millet (Eleusine coracana L.) confers tolerance against salinity and drought stress in rice. BMC Biotechnol. 16:35. https://doi.org/10.1186/s12896-016-0261-1
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. https://doi.org/10.1093/bioinformatics/btp616
Satou Y, Imai KS, Levine M, Kohara Y, Rokhsar D, Satoh N (2003) A genomewide survey of developmentally relevant genes in Ciona intestinalis - I. Genes for bHLH transcription factors. Dev Genes Evol 213:213–221. https://doi.org/10.1007/S00427-003-0319-7
Shah K, Nahakpam S (2012) Heat exposure alters the expression of SOD, POD, APX and CAT isozymes and mitigates low cadmium toxicity in seedlings of sensitive and tolerant rice cultivars. Plant Physiol Biochem 57(3):106–113. https://doi.org/10.1016/j.plaphy.2012.05.007
Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97:6896–6901. https://doi.org/10.1073/pnas.120170197
Song XM, Huang ZN, Duan WK, Ren J, Liu TK, Li Y, Hou XL (2014) Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 289:77. https://doi.org/10.1007/s00438-013-0791-3
Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu JK, Shinozaki K (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol 135(3):1697–1709. https://doi.org/10.1104/pp.104.039909
Theodoulou FL (2000) Plant ABC transporters. BBA-Biomembranes 1465(1–2):79–103. https://doi.org/10.1016/S0005-2736(00)00132-2
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25:1105–1111. https://doi.org/10.1093/bioinformatics/btp120
Ullah A, Dutta D, Fliegel L (2016) Expression and characterization of the SOS1 Arabidopsis salt tolerance protein. Mol Cell Biochem 415:133–143. https://doi.org/10.1007/s11010-016-2685-2
van de Graaf SFJ, Hoenderop JGJ, Bindels RJM (2006) Regulation of TRPV5 and TRPV6 by associated proteins. AM J Physiol Renal Physiol 290:F1295–F1302. https://doi.org/10.1152/ajprenal.00443.2005
Wang Z, Gerstein M, Snyder M (2009) RNA-seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63. https://doi.org/10.1038/nrg2484
Weinl S, Kudla J (2009) The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol 184:517–528. https://doi.org/10.1111/j.1469-8137.2009.02938.x
Wu Q, Bai X, Zhao W, Xiang D, Wan Y, Yan J, Zou L, Zhao G (2017) De novo assembly and analysis of Tartary buckwheat (Fagopyrum tataricum Gaertn.) transcriptome discloses key regulators involved in salt-stress response. Genes 8(10):255. https://doi.org/10.3390/genes8100255
Ye CY, Zhang HC, Chen JH, Xia XL, Yin WL (2009) Molecular characterization of putative vacuolar NHX-type Na+/H+ exchanger genes from the salt-resistant tree Populus euphratica. Physiol Plantarum 137(2):166–174. https://doi.org/10.1111/j.1399-3054.2009.01269.x
Ye J, Fang L, Zheng H et al (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293-297. https://doi.org/10.1093/nar/gkl031
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-Seq: accounting for selection bias. Genome Biol 11:R14. https://doi.org/10.1186/gb-2010-11-2-r14
Zhang C, Wang D, Yang C, Kong N, Zheng S, Peng Z, Nan Y, Nie T, Wang R, Ma H (2017) Genome-wide identification of the potato WRKY transcription factor family. PLoS ONE 12:e0181573. https://doi.org/10.1371/journal.pone.0181573
Zhang X, Ju HW, Chung MS, Huang P, Ahn SJ, Kim CS (2011) The R-R-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol 52:138–148. https://doi.org/10.1093/pcp/pcq180
Zhou J, Li F, Wang JL, Ma Y, Chong K, Xu YY (2009) Basic helix-loop-helix transcription factor (OrbHLH2) from wild rice enhances Arabidopsis tolerance to salt stress and osmotic stress. J Plant Physiol 166:1296–1306. https://doi.org/10.1016/j.jplph.2009.02.007
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273. https://doi.org/10.1146/annurev.arplant.53.091401.143329
Acknowledgements
The authors would like to acknowledge the financial support from the National Natural Science Foundation of China (31371552), and acknowledge Dr. Mark Ziemann of Deakin University in Australia for language editing.
Author information
Authors and Affiliations
Contributions
J-NS analyzed the relative expression level of genes and drafted the manuscript. X-HL analyzed the transcriptome data of Tartary buckwheat. Y-QW cultivated the plant and treated. H-BY designed the study and helped draft the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict exists among all the authors, and the contribution of the authors is clear and unquestionable. All of them declare that they have no conflict of interest. Therefore, all authors are allowed to publish the article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Song, JN., Liu, XH., Wang, YQ. et al. Transcriptome analysis reveals salinity responses in four Tartary buckwheat cultivars. J. Plant Biochem. Biotechnol. 30, 564–578 (2021). https://doi.org/10.1007/s13562-021-00648-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-021-00648-2