Abstract
The purpose of this study was to examine the influence of climate on the distribution and present-day genetic structure of the common vole (Microtus arvalis) and the field vole (Microtus agrestis). In this study, we used previously published data on the genetic structure (using microsatellite DNA) of the common and field vole in Central Europe and a set of climatic variables to conduct binomial generalized linear and environmental niche modeling. In terms of present-day genetic structure, climate is an important factor shaping the patterns of distribution of the identified genetic groups, with the average minimum temperature in January being a significant factor for both species. For the field vole, average annual precipitation was an important factor also and consistent with the species’ preference for wet habitats. Therefore, this study has provided indirect evidence that (1) climate can shape the genetic structure and distribution of species at both broad and local scales and (2) using genetic data and species distribution modeling can be an effective approach to establish locations of putative glacial refugia for different species in Europe and to explore their past evolutionary history.
Similar content being viewed by others
Introduction
Climate is an important factor influencing species distribution patterns worldwide (Swenson 2006; Moritz et al. 2009). Phylogeographic patterns in European mammals were significantly shaped by unfavorable climatic conditions during the last glaciation, mainly during the Last Glacial Maximum (LGM) that lasted from approximately 27.5 to 19 thousand years ago (kya) (Clark et al. 2009). Some mammalian species retreated south, to refugia located in southern Europe on the Balkan, Apennine, and Iberian peninsulas, recognized as the “traditional” southern refugia (Hewitt 1999). However, many species also survived in refugia located further north (the so called “northern refugia”), for instance in central France and the Carpathian Basin (Stewart and Lister 2001; Pazonyi 2004; Sommer and Nadachowski 2006; Stojak et al. 2016a).
The climate is changing continuously, and it could potentially modify species distributions and genetic lineages/groups over time. It is also apparent that individual species responded to changing climate differently, depending on their specific adaptations (Stewart et al. 2010). Several studies have inferred the influence of climate on both broad and fine-scale patterns of genetic structuring (McDevitt et al. 2012; Tarnowska et al. 2016). In this study, we focused on two closely related rodent species, the common vole (Microtus arvalis) and the field vole (Microtus agrestis). Although morphologically similar, they prefer different habitats, and their evolutionary history differs from each other. Both species have been well-studied in Europe with different molecular markers—mitochondrial cytochrome b sequence (Haynes et al. 2003; Heckel et al. 2005; Bužan et al. 2010; Herman and Searle 2011; Paupério et al. 2012; Martínková et al. 2013; Herman et al. 2014; Stojak et al. 2015), microsatellites (Heckel et al. 2005; Beysard et al. 2012; Stojak et al. 2016b), and Y chromosome sequences (Beysard and Heckel 2014). In Western Europe, different markers provided congruent patterns of genetic structure for both species. In Central Europe, however, contrasting patterns were found. MtDNA analysis revealed two lineages of the common vole—the Central and Eastern lineages, with the contact zone between them in north-western Poland (Stojak et al. 2015, 2016a). For the field vole, two lineages were also found in Central Europe—the Western and Central-European lineages with the contact zone in south-western Poland (Herman et al. 2014). Based on microsatellite DNA, two genetic groups were found for each species, named according to their geographic distribution—the Western and Eastern groups (Stojak et al. 2016b; Fig. S1), with these patterns being generally similar between both species. Moreover, the distribution of mtDNA lineages and genetic groups based on microsatellites was not congruent within either species. It was proposed that the genetic structure observed in the microsatellite data could have been influenced by contemporary environmental conditions (Stojak et al. 2016a).
Given the distribution of genetic groups identified using microsatellite markers observed for the common vole and field vole in Central Europe (Stojak et al. 2016a, b), the purpose of this study was to examine if climatic conditions could have had an influence on the present-day genetic structure of both species. In addition, we used environmental niche modeling to examine the relationship between present-day species distributions and putative distribution of these two species during the LGM.
In this study, we put forward the following hypotheses: (1) the observed contemporary genetic structure based on microsatellite loci and distribution of the common vole and field vole in Central Europe was affected by climatic conditions; (2) in spite of different habitat preferences, these two vole species could respond in a similar way to recent climate changes; and (3) species distribution modeling based on climatic conditions may be used to indicate the locations of putative glacial refugia of common and field voles and to explore their past evolutionary history.
Methods
In this study, we used data on microsatellite markers in the common vole and field vole published in previous studies (Stojak et al. 2016a, b), including the distribution of the genetic groups of these species in Central Europe (see Tables S1 and S2; Fig. S1). Genetic groups were identified using Bayesian clustering analysis of the microsatellite data conducted in STRUCTURE v. 2.3.3 (Pritchard et al. 2000). The number of genetic clusters for these two species was distinguished across Central Europe, testing K values from 1 to 10 using ten independent runs of 500,000 generations (including 100,000 generations of burn-in) under the admixture model (Stojak et al. 2016b). The optimal value of K was determined using the Evanno method (ΔK, Evanno et al. 2005). For the common vole, 311 individuals from 61 locations were genotyped using eight microsatellite loci (FST = 0.040), while for the field vole, 190 individuals from 23 locations were genotyped using 13 loci (FST = 0.047) (Stojak et al. 2016a, b). Assignment of individuals to a particular genetic cluster was carried out according to the criteria of Vähä and Primmer (2006) with an assignment probability of q ≥ 0.8. Admixed individuals (with an assignment probability of 0.2 < q < 0.8) were observed only in a few populations of both species at a low level (see Stojak et al. 2016a, b), and these individuals were excluded from further analyses. The distribution of populations belonging to the Eastern and Western groups (and populations with individuals from both genetic groups present) is presented in Fig. S1. The exact number of individuals belonging to each genetic group is given in Tables S1 and S2.
To test if climatic conditions could explain the spatial distribution of the genetic groups in the common and field voles in Central Europe, we used binomial generalized linear modeling (GLM; Zuur et al. 2009). GLMs are an effective tool for investigating associations between environmental variables and genetic groups/clusters/lineages (McDevitt et al. 2012; Theodoridis et al. 2017; Yannic et al. 2014). For each rodent population, we calculated the set of climatic variables at a 1 km2 resolution, including minimum temperature in January, maximum temperature in July, average annual temperature, and average annual precipitation (combined rainfall and snow cover) based on the WorldClim database (version 2; 1970–2000; http://www.worldclim.org/) using ArcMap GIS software by ESRI (version 10.3.1; ArcGIS Desktop: Release 10.3 Redlands, CA: Environmental Systems Research Institute 2015). These four climatic variables were chosen because of the existence of a strong west-east precipitation and seasonality gradients in Europe.
The average minimum temperature in January ranged from − 1.1 °C to − 7.7 °C in the common vole populations and from − 1.4 °C to − 5.3 °C in the field vole. The average maximum temperature in July varied from 18.4 °C to 25.7 °C for the common vole and from 18.6 °C to 22.3 °C for the field vole. The average annual temperatures were between 4.7 °C and 11.4 °C for the common vole and between 5.9 °C and 8.9 °C for the field vole. For the common vole, the average annual precipitation ranged from 40.4 mm to 122.1 mm, and from 41.5 mm to 76.7 mm for the field vole (Tables S1 and S2). For the dependent binomial variable, we used an assignment to either of the two genetic groups (Eastern or Western) from previously published papers, established using the STRUCTURE analysis described above (for further details, see Stojak et al. 2016a, 2016b; Fig. S1). In further analyses, we used three non-correlated variables (R < 0.4): minimum temperature in January, maximum temperature in July, and average annual precipitation at the location of sampled populations.
The genetic structuring indicating different climatic preferences of the genetic groups could potentially be the result of a different refugial origin for each group. In our study area, the Carpathian refugium is present (Stewart and Lister 2001; Pazonyi 2004; Sommer and Nadachowski 2006). Stojak et al. (2015, 2016a) suggested that the Eastern mtDNA lineage of the common vole could have survived the LGM in the Carpathian refugium and re-colonized Eastern and Central Europe from this area. Therefore, in the case of this species, we additionally checked for each population if there was any association between the distance from the potential refugial area in the Carpathians and being assigned to the Eastern or Western genetic group (using GLM analysis). We assume such an association of any genetic group could potentially suggest that the observed genetic structure was the result of originating of each genetic group from two different glacial refugia. The exact location of the Carpathian refugium is unknown, but fossil data suggests that it occupied what is present-day north-eastern Hungary (Pazonyi 2004; Sommer and Nadachowski 2006). Therefore, we have arbitrarily chosen a random location in this area as a point representing the Carpathian refugium (see Fig. S1a, based on Pazonyi 2004). We adopted a similar approach to McDevitt et al. (2012) by measuring the Euclidean distance between each location of the common vole populations (for the Eastern and Western genetic group separately) and location point represented refugial area. Calculated distance from the refugium is given in Table S1.
Global models (for the common and field vole) were ranked with the Akaike Information Criterion (AIC) with a second-order correction for small sample size (AICc) (Burnham and Anderson 2002; Arnold 2010; Richards et al. 2011) available within the dredge function (“MuMIn” package; Bartoń 2015). AIC allows us to estimate the relative quality of statistical models for a given set of data, where the preferred model is the one with the minimum AIC value. We have chosen the best model based on a visual inspection of the differences in ΔAICC. All statistical analyses were done in R (R Development Core Team 2018).
For the climatic factors which best explained the current spatial distribution of the genetic groups for the common vole and field vole populations, we prepared spatial interpolation of values from all locations (Tables S1 and S2) using ArcGIS Geostatistical Analyst. Interpolation was based on inverse distance weight model (IDW, power = 1, based on 12 neighbors) which determines cell values using a linearly weighted combination of a set of measured values of sample points, where the weight is a function of inverse distance from the output cell location.
According to the hypothesis that climate has shaped species distribution across their entire ranges, we aimed to test how temperature and climate humidity influenced the distribution of the common vole and field vole, species with different habitat preferences, in the past. Such predictions could deliver new insights into how these two small mammal species were distributed in the past and to test if the glacial refugia locations suggested by the genetic (based on previous phylogeographic studies using mtDNA) and climatic data are congruent. In order to achieve this, we performed species distribution modeling (SDM) for the two species, based on a mathematical representation of already described environmental preferences of the species, which depend on a number of factors, such as climatic factors, barriers to dispersal, geologic history, or biotic interactions (Elith and Leathwick 2009; Morin and Thuiller 2009). We used MAXENT 3.3.3k software exploiting the maximum enthropy method (Phillips et al. 2006; Elith et al. 2011). MAXENT is a modeling tool characterized by high predictive accuracy. It allows us to combine information about species occurrence with present-day climate data to predict how species niches and suitable climatic conditions were distributed in the past.
Distribution data of common and field voles in Eurasia were downloaded from the IUCN Redlist (Yigit et al. 2016; Kryštufek et al. 2016) website, and the whole range area covered by each species was treated as a polygon and transformed into point data with 2.5 min spatial resolution in ArcGIS. To build the models of distribution, we used the same three non-correlated bioclimatic variables as for the GLM, representing the (1) present (1960–1990) and (2) LGM (21 kya) conditions. Environmental data were downloaded from WorldClim database with 2.5 min spatial resolution and converted into ASCII raster grids in ArcGIS. The present distribution model was projected to the LGM models. For the LGM, we downloaded climatic data from WorldClim database for two atmospheric circulation models constructed in the Paleoclimate Modeling Intercomparison Project Phase II (Braconnot et al. 2007): Model for Interdisciplinary Research on Climate (MIROC; Hasumi and Emori 2004) and Community Climate System Model (CCSM; Collins et al. 2006).
Our model parameters were as follows: a convergence threshold of 10−5 with 500 iterations and 100,000 background points, auto features, a regularization multiplier of 1, random seed, response curve, a jackknife test of variable importance, and all environmental data treated as continuous layers. Distribution data were randomly split into training data (70%) and test data (30%; Pearson 2007). All duplicate records were removed. Maximum test sensitivity plus specificity threshold was used, which maximizes the cases where the model erroneously assigns unsuitable habitat and misses suitable habitat. The MESS and clamping analyses were performed during projection to identify areas of low reliability of predictions. Each model was replicated ten times with bootstrapping, and the final model was chosen on the basis of average values obtained from all ten replicates. The logistic output format was used, and the pictures of predictions were prepared with colors indicating predicted probability that conditions were or were not suitable for the species: red color indicated high probability, yellow indicated typical conditions where species could be found, and lighter shades of blue indicated low probability of suitable conditions. Models were evaluated by calculating the area under the curve (AUC; where AUC values under 0.7 indicate poor model performance, and values higher than 0.9 indicate excellent model performance; Fielding and Bell 1997), and species distribution predictions were evaluated using omission error (false negative predictions; Fielding and Bell 1997).
Results
For the common vole, among the different combinations of the considered submodels of the GLM aimed at assessing the effect of the climatic factors on the assignment to one of the genetic groups, the model with average minimum temperature in January was selected as the single best model (Table 1). This model was the simplest and had the lowest AICc scores and indicated that the chance of a population to being assigned to the Eastern group increased with decreasing minimum temperature in January (z-value = − 3.33, p = 0.001). Populations from the coldest locations (< − 4 °C) had 80% probability to be assigned to the Eastern group, while populations from warmer locations (> − 2 °C) had less than a 10% chance to belong to the Eastern group (Figs. 1a and 2). The addition of other variables in the second-, third-, and fourth-ranked submodels did not substantially reduce model deviance (Table S3). Therefore, we selected only the top-ranked submodel as a single best model (Arnold 2010; Table 1). The impact of distance from the refugium on the genetic structure in this species was negligible (Tables 1 and S3).
The probability of an individual belonging to the Eastern genetic group of Microtus arvalis (a) and Microtus agrestis (b, c) in relation to the average minimum temperature in January (a, b) and average annual precipitation (c) of the sampling location. The gray bars on the top represent the Eastern genetic groups, while the gray bars on the bottom represent the Western genetic group. The “Frequency” axis showed the number of individuals belonging to each group found on the area with certain temperature presented on x axis. The line is a predicted value based on the generalized linear model
The interpolation of average minimum temperature values in January from sampled locations and the distribution of the two genetic groups of Microtus arvalis in Central Europe. The Eastern genetic group is marked by black triangles, and the Western group is marked by gray circles. Populations with the majority of individuals assigned to the Eastern group are marked by white triangles. Populations with the majority of individuals assigned to the Western group are marked by white circles (see Fig. S1a; Table S1)
For the field vole, two top-ranking models of the GLM were chosen with the lowest AICc (Table 2). They indicated that average minimum temperature in January (z-value = − 2.39, p = 0.02) and average annual precipitation (z-value = − 2.21, p = 0.03) had significant negative effects on the probability of being assigned to the Eastern group. Maximum temperature in July did not significantly influence the genetic assignment in the field vole populations. Populations from the coldest locations (< − 4 °C) had an 80% probability of being assigned to the Eastern group. The lowest chance (below 5%) had field vole populations occurring in areas where minimum temperature in January exceeded − 2 °C (Fig. 1b). The assignment probability to the Eastern group decreased from 85 to 15% when average annual precipitation level increased from 45 to 55 mm (Figs. 1c and 3).
The interpolation of average minimum temperature in January (a) and average annual precipitation (b) values from sampled locations and the distribution of the two genetic groups of Microtus agrestis in Poland. The Eastern genetic group is marked by black triangles, and the Western group is marked by gray circles. Populations with the majority of individuals assigned to the Western group are marked by white circles (see Fig. S1b; Table S2)
The species distribution modeling for the common and field voles showed that all predictive models were statistically significant, with high AUC values and low omission rates (Table 3). In general, the current distribution obtained by modeling in MAXENT was consistent with the actual ranges of the common vole and the field vole (Figs. 4 and 5). Notwithstanding, in the case of the common vole, there were some inconsistencies between the IUCN current species distribution and that obtained in MAXENT. The model of the current distribution obtained in our analyses implied that it was likely for the common vole to be found on the British Isles (Fig. 4). However, the common vole is present only on the Orkney archipelago where it was introduced by humans (Martínková et al. 2013). Present-day Great Britain is inhabited by the field vole which was predicted in the obtained model for this species (Fig. 5). The opposite situation was observed in area of the Altai Mountains, which has not been identified as potentially suitable for the common vole (Fig. 4). However, this area is currently inhabited by the subspecies M. arvalis obscurus, also known as the Altai vole (Meyer et al. 1997).
Predicted current distribution of Microtus arvalis (a) and during LGM under MIROC (b) and CCSM models (c). Red color indicates high probability of suitable conditions, yellow indicates typical conditions where species could be found, and lighter shades of blue indicates low probability. Continuous black line represents actual range of Microtus arvalis according to IUCN
Predicted current distribution of Microtus agrestis (a) and during LGM under MIROC (b) and CCSM models (c). Red color indicates high probability of suitable conditions, yellow indicates typical conditions where species could be found, and lighter shades of blue indicates low probability. Continuous black line represents actual range of Microtus agrestis according to IUCN
Potential distribution areas for the common vole during the LGM (glacial refugia) were concentrated on three Mediterranean peninsulas (Apennine, Iberian, and Balkan), in the Carpathian Basin, Caucasus, and Crimea (Fig. 4). Highly suitable areas (≥ 0.7 suitability) represented not more than 5% of the area (Fig. 4). For the field vole, potential distribution areas during the LGM (glacial refugia) were concentrated on the Iberian Peninsula, southern part of Apennine Peninsula, Romania and Bulgaria, Caucasus, and near Bosphorus (Fig. 5). Highly suitable areas (≥ 0.7 suitability) represented not more than 10% of the area (Fig. 5).
Discussion
In this study, we assumed that different climatic variables present across Central Europe could potentially influence the genetic structure and the distribution of genetic groups identified for the common vole and field vole using microsatellite DNA. The differences in temperature and precipitation amplitudes across Central Europe are not drastic; however, the GLM analyses suggested that distinct differentiation of climatic conditions in the eastern and western parts of our study area is a significant for the observed genetic structure in the common and field voles’ populations.
The distribution of the genetic groups of the common vole was positively correlated with the average minimum temperature of January only, while the distribution of the field vole groups was positively correlated with two factors—average minimum temperature of January and average annual precipitation (Table 2). For the field vole, the influence of precipitation is consistent with the habitat preferences of the species as it is found in wet habitats such as upland heaths, marshes, peat bogs, and river banks (Mitchell-Jones et al. 1999). The common vole on the other hand tends to be found in open habitats such as pastures, forest steppe, or agricultural areas. Therefore, such results are consistent with the environmental requirements of these rodents (Mitchell-Jones et al. 1999).
It suggests that voles belonging to the Eastern genetic groups of both species could be better adapted to the colder continental climate, while the Western genetic groups of these species are associated with a somewhat milder climate. Such adaptations have not been experimentally tested, and associations mentioned above remain contentious. However, results presented in this study might indicate that strong west-east precipitation and seasonality gradients in Europe could influence gene flow shaping the contemporary genetic structure of different small mammal species. Although these environmental gradients are well-known in this region, its relationship with patterns of contemporary genetic structuring in two closely related species is not. This study suggests that small mammals, even those that differ in their ecological and habitat preferences, are highly susceptible to cold temperatures in winter, and different mammal species present on the same region could react in a similar way for the same climatic conditions. For voles specifically (including the two species that have been studied here), climate has emerged as a key extrinsic factor influencing population cycles associated with a reduction in winter population growth (Cornulier et al. 2013). The effects of these cycles on patterns of gene flow in voles have only been studied at local spatial and short-term temporal scales (Gauffre et al. 2014), but climate has been demonstrated to be a key driver in determining the distribution of genetic clusters within other mammalian species, even over relatively short periods of time in a rapidly warming world (Rubidge et al. 2012).
Similar studies as performed here were conducted for the weasel and bank vole using mitochondrial cytochrome b sequences. It has been proposed that the Carpathian lineage of weasel had an advantage over other lineages (for instance the Balkan and Eastern lineages) in terms of being better adapted to a colder, more severe climate (McDevitt et al. 2012). Moreover, the distribution of the Carpathian lineage of bank vole was positively correlated with the mean temperature of July and the occurrence of plant species associated with the Carpathian refugium (Tarnowska et al. 2016). Both weasel and bank vole are proposed to have occupied the Carpathian refugium during the LGM based on both fossil and genetic data. In the case of the common vole and field vole analyzed in this study, only the common vole likely occupying this refugium during the LGM (Herman et al. 2014; Stojak et al. 2015, 2016a). While originating from this particular refugium is reflected in the distribution of the mitochondrial DNA lineages in weasels, bank voles, and common voles in this region (Wójcik et al. 2010; McDevitt et al. 2012; Stojak et al. 2016a; Tarnowska et al. 2016), the best selected model in the GLM analysis did not include distance from the Carpathian refugium, showing no association between refugial origin and the observed distribution of genetic groups in the common vole based on microsatellite data. Thus, it can be inferred that refugial origin was not a crucial factor in shaping the contemporary genetic structure of this species in Central and Eastern Europe (Tables 1 and S3) and that the patterns observed might have been indirectly driven by other factors, e.g., by more recent changes in the environment in these species.
The species distribution modeling for the common and field voles seems to confirm the locations of the glacial refugia proposed for these two species by different authors (Hewitt 1999; Jaarola and Searle 2002; Haynes et al. 2003; Marková 2011; Paupério et al. 2012; Martínková et al. 2013; Stojak et al. 2016a, b). Both models (MIROC and CCSM) of the common vole distribution during the LGM suggested a plausible refugial area in the Carpathian region (Fig. 4). As it was already mentioned, two phylogenetic lineages of the common vole can be found in Central Europe, and previous research indicated that the Eastern mtDNA lineage originated from the Carpathian refugium (Stojak et al. 2015, 2016a). During the LGM, this region was covered by arctic-like tundra, and the climate was more severe than in southern Europe. Therefore, individuals from the Eastern mtDNA lineage might have been better adapted to the cold climatic conditions than individuals from other lineages of the common vole which survived the LGM in the three Mediterranean peninsulas (as in the case of the weasels; McDevitt et al. 2012). The origin of the Central mtDNA lineage of the common vole has not been revealed yet; however, modeling results might confirm previous hypothesis that it could be located in the northern part of Apennine Peninsula, in close proximity to the Alps (Fig. 4). The existence of a glacial refugium in this locality is additionally indicated by the distribution of the Central mtDNA lineage in Europe, the location of the alpine region, its distance from Central Europe, and the contact zone between the Central and Eastern mtDNA lineages of this species in Poland. The location of suitable habitat in close proximity to the Alps is also concordant with the hypothesis of transalpine colonization which assumes the crossing of mountain ranges at several locations after the LGM and populating various sites characterized by suitable ecological conditions (Braaker and Heckel 2009).
The SDM of the current distribution of the common vole also implied that it was likely for this species to be found in the British Isles, while it was highly unlikely for it to be found in the Altai Mountains (Fig. 4). Such inconsistencies are a bias caused by disregarding in SDM projections the geographical barriers (Panzacchi et al. 2015) and a result of the modeling assumptions according to which species of interest is in equilibrium and always inhabits highly suitable habitats. However, the real process of species expanding/retracting their distribution range is more complicated and difficult to examine in silico. Not only climate and habitat suitability need to be taken into account but also unstable and difficult to measure factors such as interspecific competition, niche availability, human impacts, barriers, and capacity for adaptations.
In the case of the field vole, previous research based on fossil records indicated western Iberia, north-eastern Spain, southern France, and northern Italy as refugial areas for this species (Paupério et al. 2012). Both models (MIROC and CCSM) obtained in this study seem to be congruent with these data (Fig. 5). Additionally, Jaarola and Searle (2002) proposed refugia located further to the east, such as the Carpathians and Urals. However, no fossil data of the field vole confirming this has been found (Pazonyi 2004; Sommer and Nadachowski 2006), and neither do the SDM models suggest it (Fig. 5). According to Pazonyi (2004), this rodent was not present in the Carpathian Basin until after the Younger Dryas glaciation (12.9–11.7 kya). It is perhaps not surprising for a species which is less tolerant to dry climate than its sibling species, the common vole. Under arid and cold glacial conditions, the field vole may have survived in small populations in areas where the environment and climate were tolerable and the competition level was low, for instance in the Balkans (Fig. 5).
We are aware that there are still many shortcomings in methodology used for species distribution modeling and revealing their evolutionary history. It is difficult to include all the probable factors that contribute to the complex and complicated dynamics present in the environment during the last ca. 25 kya (i.e., local differences in climatic conditions, geographical and anthropogenic barriers, interspecific competition). Nevertheless, we believe that results obtained in this study allow us to bring new inferences in potential mechanisms shaping worldwide biodiversity. Inconsistencies between actual (based on IUCN) and predicted past species ranges shown in our study show that multiple approaches are required to obtain reliable results (genetics, SDM, fossil data).
This study has provided indirect evidence that climate is shaping the contemporary genetic structure and the distribution of the common vole and field vole at both broad and local scales. Moreover, knowledge on habitat preferences could be used in predicting where species of interest could survive during the last glaciation, how it re-colonized currently inhabited areas, and how climate change could potentially affect its distribution in the future (Ikeda et al. 2017; Theodoridis et al. 2017). Such predictions are highly significant for instance in conservation biology, allowing to take quick action to protect species and their habitats from extinction. SDM could also deliver new insights into the biology, ecology, and biogeography of different species of flora and fauna in the past, allowing us to describe community compositions (Fløjgaard et al. 2009; Gür et al. 2018). Additionally, this study proved that it is possible to successfully predict species distribution in the past on the basis of current climatic conditions, which helps us to understand the dynamics of environments in the past and how they have shaped present-day distributions and patterns of genetic structure/groupings.
References
Arnold TW (2010) Uninformative parameters and model selection using Akaike’s Information Criterion. J Wildlife Manage 74:1175–1178
Bartoń K (2015) MuMIn: multi-model inference (R package version 1.15.1). Available at: https://cran.r-project.org/web/packages/MuMIn/index.html
Beysard M, Heckel G (2014) Structure and dynamics of hybrid zones at different stages of speciation in the common vole (Microtus arvalis). Mol Ecol 23:673–687
Beysard M, Perrin N, Jaarola M, Heckel G, Vogel P (2012) Asymetric and differential gene introgression at a contact zone between two highly divergent lineages of field voles (Microtus agrestis). J Evol Biol 24:400–408
Braaker S, Heckel G (2009) Transalpine colonization and partial phylogeographic erosion by dispersal in the common vole (Microtus arvalis). Mol Ecol 18:2518–2531
Braconnot P, Otto-Bliesner B, Harrison S, Joussaume S, Peterchmitt JY, Abe-Ouchi A, Crucifix M, Driesschaert E, Fichefet T, Hewitt CD, Kageyama M, Kitoch A, Laîné A, Loutre MF, Marti O, Merkel U, Ramstein G, Valdes P, Weber SL, Yu YZY (2007) Results of PMIP2 coupled simulations of the mid-Holocene and last glacial maximum – part 1: experiments and large-scale features. Clim Past 3:261–277
Burnham KP, Anderson DR (2002) Model selection and multimodel inference: a practical information-theoretic approach. Springer, New York
Bužan EV, Förster DW, Searle JB, Kryštufek B (2010) A new cytochrome b phylogroup of the common vole Microtus arvalis endemic to the Balkans and its implications for the evolutionary history of the species. Biol J Linn Soc 100:788–796
Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B, Mitrovica JX, Hostetler SW, McCabe AM (2009) The last glacial maximum. Science 325:710–714
Collins WD, Bitz CM, Blackmon ML, Bonan GB, Bretherton CS, Carton JA, Chang P, Doney SC, Hack JJ, Henderson TB, Kiehl JT, Large WG, McKenna DS, Santer BD, Smith RD (2006) The Community Climate System Model version 3 (CCSM3). J Clim 19:2122–2143
Cornulier T, Yoccoz NG, Bretagnolle V, Brommer JE, Butet A, Ecke F, Elston DA, Framstad E, Henttonen H, Hörnfeldt B, Huitu O, Imholt C, Ims RA, Jacob J, Jędrzejewska B, Millon A, Petty SJ, Pietiäinen H, Tkadlec E, Zub K, Lambin X (2013) Europe-wide dampening of population cycles in keystone herbivores. Science 340:63–66
Elith J, Leathwick JR (2009) Species distribution models: ecological explanation and prediction across space and time. Annu Rev Ecol Evol Syst 40:677–697
Elith J, Phillips SJ, Hastie T, Dudík M, Chee YE, Yates CJ (2011) A statistical explanation of MaxEnt for ecologist. Divers Distributions 17:43–57
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fielding AH, Bell JF (1997) A review of methods for the assessment of prediction errors in conservation presence/ absence models. Environ Conserv 24:38–49
Fløjgaard C, Normand S, Skov F, Svenning JC (2009) Ice age distributions of European small mammals: insights from species distribution modelling. J Biogeogr 36:1152–1163
Gauffre B, Berthier K, Inchausti P, Chaval Y, Bretagnolle V, Cosson JF (2014) Short-term variations in gene flow related to cyclic density fluctuations in the common vole. Mol Ecol 23:3214–3225
Gür H, Perktaş U, Gür MK (2018) Do climate-driven altitudinal range shift explain the intraspecific diversification of a narrow ranging montane mammal, Taurus ground squirrels? Mammal Res 63:197–211
Hasumi H, Emori S (2004) K-1 coupled model (MIROC) description. K-1 technical report. University of Tokyo, Tokyo, p 1
Haynes S, Jaarola M, Searle JB (2003) Phylogeography of the common vole Microtus arvalis with particular emphasis on the colonization of the Orkney archipelago. Mol Ecol 12:951–956
Heckel G, Burri R, Fink S, Desmet JF, Excoffier L (2005) Genetic structure and colonization processes in European populations of the common vole Microtus arvalis. Evolution 59:2231–2242
Herman JS, Searle JB (2011) Post-glacial partitioning of mitochondrial genetic variation in the field vole. Proc R Soc Lond 278:3601–3607
Herman JS, McDevitt AD, Kawałko A, Jaarola M, Wójcik JM, Searle JB (2014) Land-bridge calibration of molecular clocks and the post-glacial colonization of Scandinavia by the Eurasian field vole Microtus agrestis. PLoS One 9:e103949
Hewitt GM (1999) Post-glacial re-colonization of European biota. Biol J Linn Soc 68:87–112
Ikeda DH, Max TL, Allan GJ, Lau MK, Shuster SM, Whitham TG (2017) Genetically informed ecological niche models improve climate change predictions. Glob Change Biol 23:164–176
Jaarola M, Searle JB (2002) Phylogeography of field voles (Microtus agrestis) in Eurasia inferred from mitochondrial DNA sequences. Mol Ecol 11:2613–2621
Kryštufek B, Vohralík V, Zima J, Zagorodnyuk I (2016) Microtus agrestis. The IUCN red list of threatened species, e.T13426A115112050
Marková AK (2011) Small mammals from Palaeolithic sites of the Crimea. Quat Int 231:22–27
Martínková N, Barnett R, Cucchi T, Struchen R, Pascal M, Pascal M, Fischer MC, Higham T, Brace S, Ho SYW, Quéré JP, O’Higgins P, Excoffier L, Heckel G, Hoelzel AR, Dobney KM, Searle JB (2013) Divergent evolutionary processes associated with colonization of offshore islands. Mol Ecol 22:5205–5220
McDevitt AD, Zub K, Kawałko A, Oliver MK, Herman JS, Wójcik JM (2012) Climate and refugial origin influence the mitochondrial lineage distribution of weasels Mustela nivalis in a phylogeographic suture zone. Biol J Linn Soc 106:57–69
Meyer MN, Golenishchev FN, Bulatova NS, Artobolevsky GV (1997) On distribution of two Microtus arvalis chromosomal forms in European Russia. Zoologiskii Zhurnal 76:487–493 [in Russian]
Mitchell-Jones AJ, Amori G, Bogdanowicz W, Kryštufek B, Reijnders PJH, Spitzenberger F, Stubbe M, Thissen JBM, Vohralik V, Zima J (1999) The atlas of European mammals. Poyser Natural History, London
Morin X, Thuiller W (2009) Comparing niche- and process-based models to reduce prediction uncertainty in species range shifts under climate change. Ecology 90:1301–1313
Moritz C, Hoskin CJ, MacKenzie JB, Phillipis BL, Tonione M, Silva N, VanDerWal J, Williams SE, Graham CH (2009) Identification and dynamics of a cryptic suture zone in tropical rainforest. Proc R Soc Lond 276:1235–1244
Panzacchi M, Moorter BV, Strand O, Loe LE, Reimers E (2015) Searching for the fundamental niche using individual-based habitat selection modelling across populations. Ecography 38:659–669
Paupério J, Herman JS, Melo-Ferreira J, Jaarola M, Alves PC, Searle JB (2012) Cryptic speciation in the field vole: a multilocus approach confirms three highly divergent lineages in Eurasia. Mol Ecol 21:6015–6032
Pazonyi P (2004) Mammalian ecosystem dynamics in the Carpathian Basin during the last 27000 years. Palaeogeogr Palaeoclimatol Palaeoecol 212:295–314
Pearson RG (2007) Species’ distribution modelling for conservation educators and practitioners synthesis. American Museum of Natural History [online document]. URL http://biodiversityinformatics.amnh.org/filesSpeciesDistModellingSYN_1-16-08.pdf
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259
Pritchard JK, Stephens M, Donelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Richards SA, Whittingham MJ, Stephens PA (2011) Model selection and model averaging in behavioural ecology: the utility of the IT-AIC framework. Behav Ecol Sociobiol 65:77–89
RStudio Team (2018) RStudio: integrated development for R (Version 1.1.453). Computer software, Boston, MA
Rubidge EM, Patton JL, Lim M, Burton AC, Brashares JS, Moritz C (2012) Climate-induced range contraction drives genetic erosion in an alpine mammal. Nat Clim Chang 2:285–288
Sommer RS, Nadachowski A (2006) Glacial refugia of mammals in Europe: evidence from fossil records. Mammal Rev 36:251–265
Stewart JR, Lister AM (2001) Cryptic northern refugia and the origins of the modern biota. Trends Ecol Evol 16:608–613
Stewart JR, Lister AM, Barnes I, Dalén L (2010) Refugia revisited: individualistic responses of species in space and time. Proc R Soc Lond 277:661–671
Stojak J, McDevitt AD, Herman JS, Searle JB, Wójcik JM (2015) Post-glacial colonization of eastern Europe from the Carpathian refugium: evidence from mitochondrial DNA of the common vole Microtus arvalis. Biol J Linn Soc 115:927–939
Stojak J, McDevitt AD, Herman JS, Kryštufek B, Uhlíková J, Purger JJ, Lavrenchenko LA, Searle JB, Wójcik JM (2016a) Between the Balkans and the Baltic: phylogeography of a common vole mitochondrial DNA lineage limited to Central Europe. PLoS One 11(12):e0168621
Stojak J, Wójcik JM, Ruczyńska I, Searle JB, McDevitt AD (2016b) Contrasting and congruent patterns of genetic structuring in two Microtus vole species using museum specimens. Mammal Res 61:141–152
Swenson NG (2006) GIS-based niche models reveal unifying climatic mechanisms that maintain the location of avian hybrid zones in a North American suture zone. J Evol Biol 19:717–725
Tarnowska E, Niedziałkowska M, Gerc J, Korbut Z, Górny M, Jędrzejewska B (2016) Spatial distribution of the Carpathian and eastern mtDNA lineages of the bank vole in their contact zone relates to environmental conditions. Biol J Linn Soc 119:732–744
Theodoridis S, Patsiou TS, Randin C, Conti E (2017) Forecasting range shifts of a cold-adapted species under climate change: are genomic and ecological diversity within species crucial for future resilience? Ecography 41:1–13. https://doi.org/10.1111/ecog.03346
Vähä JP, Primmer CR (2006) Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol Ecol 15:63–72
Wójcik JM, Kawałko A, Marková S, Searle JB, Kotlík P (2010) Phylogeographic signatures of northward post-glacial colonization from high-latitude refugia: a case study of bank voles using museum specimens. J Zool 281:249–262
Yannic G, Pellissier L, Ortego J, Lecomte N, Couturier S, Cuyler C, Dussault C, Hundertmark KJ, Irvine RJ, Jenkins DA, Kolpashikov L, Mager K, Musiani M, Parker KL, Røed KH, Sipko T, Pórisson SG, Weckworth BV, Guisan A, Bernatchez L, Côté SD (2014) Genetic diversity in caribou linked to past and future climate change. Nat Clim Chang 4:132–137
Yigit N, Hutterer R, Kryštufek B, Amori G (2016) Microtus arvalis. The IUCN red list of threatened species 2016, e.T13488A22351133
Zuur AF, Ieno EN, Walker NJ, Saveliev AA, Smith GM (2009) Mixed effects models and extensions in ecology with R. Springer, New York
Acknowledgements
ADM received a Santander Universities award for travel to Poland. The authors thank the anonymous reviewers for their helpful comments which allowed us to improve this manuscript significantly.
Funding
The present study was financed by the National Science Centre in Poland (UMO-2013/09/N/NZ8/03205 to JS).
Author information
Authors and Affiliations
Contributions
JS, ADM, and JMW conceived the ideas. JS, JMW, TB, and MG analyzed data. JS, JMW, ADM, TB, and MG wrote the paper.
Corresponding authors
Additional information
Communicated by: Cino Pertoldi
Electronic supplementary material
Fig. S1
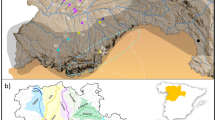
The genetic structure of Microtus arvalis (a) and Microtus agrestis (b). The genetic groups were determined in STRUCTURE, based on microsatellite DNA, described in the previous study by Stojak et al. 2016ab. The Eastern genetic group is marked by black triangles and the Western group is marked by gray circles. Populations with the majority of individuals assigned to the Eastern group are marked by white triangles. Populations with the majority of individuals assigned to the Western group are marked by white circles. The numbers referring to particular populations, including the exact number of individuals belonging to each genetic group are given in Tables S1 and S2. The potential refugial area for the common vole is marked by yellow star (a). (PNG 512 kb)
Table S1
(DOC 103 kb)
Table S2
(DOC 51 kb)
Table S3
(DOCX 11 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Stojak, J., Borowik, T., Górny, M. et al. Climatic influences on the genetic structure and distribution of the common vole and field vole in Europe. Mamm Res 64, 19–29 (2019). https://doi.org/10.1007/s13364-018-0395-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13364-018-0395-8