Abstract
The small ubiquitin-like modifier (SUMO) is a member of the family of ubiquitin-like modifiers (UBLs) and is involved in important cellular processes, including DNA damage response, meiosis and cellular trafficking. The large-scale identification of SUMO peptides in a site-specific manner is challenging not only because of the low abundance and dynamic nature of this modification, but also due to the branched structure of the corresponding peptides that further complicate their identification using conventional search engines. Here, we exploited the unusual structure of SUMO peptides to facilitate their separation by high-field asymmetric waveform ion mobility spectrometry (FAIMS) and increase the coverage of SUMO proteome analysis. Upon trypsin digestion, branched peptides contain a SUMO remnant side chain and predominantly form triply protonated ions that facilitate their gas-phase separation using FAIMS. We evaluated the mobility characteristics of synthetic SUMO peptides and further demonstrated the application of FAIMS to profile the changes in protein SUMOylation of HEK293 cells following heat shock, a condition known to affect this modification. FAIMS typically provided a 10-fold improvement of detection limit of SUMO peptides, and enabled a 36% increase in SUMO proteome coverage compared to the same LC-MS/MS analyses performed without FAIMS.

ᅟ
Similar content being viewed by others
Introduction
The small ubiquitin-like modifier (SUMO) is a member of the larger family of ubiquitin-like modifiers (UBLs) that share structural and evolutionary relationships with ubiquitin [1, 2]. Protein SUMOylation is involved in numerous cellular pathways in both the nucleus and the cytoplasm including DNA replication, DNA damage response, cell division, repression of transcription, nuclear trafficking, and receptor internalization at the plasma membrane [3,4,5]. This modification is conjugated to its substrate lysine amino group via an enzyme cascade that resembles ubiquitylation and comprises E1 activating, E2 conjugating, and E3 ligating enzymes [6, 7]. SUMOylated lysine residues were first reported to lie in the consensus motif ψKxE/D where ψ is a large hydrophobic residue, K is the modified lysine, and x is any residue preceding the glutamate (E) or aspartate (D) residues. However, other consensus sequences were reported such as a phospho-dependent sequence, reverse consensus, and non-consensus regions [7]. SUMO is also one of few UBLs that exist as multiple paralogs in human cells and SUMO1–3 are ubiquitously expressed in all tissues. While SUMO2 and SUMO3 shared 97% sequence identity, they are only ~ 50% identical to SUMO1 [8]. SUMO-2/3 has the ability to form polySUMO chains by covalent binding via the lysine residue at the N terminus consensus motif KxE/D [9]. On the other hand, SUMO-1 is unable to polymerize readily since it lacks the consensus site and is thought to act as a polySUMO chain terminator.
The identification of SUMOylation sites by mass spectrometry (MS) is challenging due to the relatively low abundance of SUMOylated proteins, the highly dynamic nature of the modification, and the relatively long SUMO remnant appended on lysine residues following tryptic digestion (e.g., up to 32 amino acids for SUMO2/3) that further complicate the interpretation of MS/MS spectra. To circumvent those issues, different approaches that used affinity purification of SUMO mutant proteins were proposed. Our group generated a functional 6xHis-SUMO3-Q87R/Q88N mutant [10] that enables the enrichment of SUMOylated proteins on a nickel-nitrilotriacetic acid (Ni-NTA) column. A five amino acid SUMO remnant is left on the substrate upon trypsin digestion, and the corresponding peptides can be immunoprecipitated using an antibody that recognizes the NQTGG epitope [11,12,13,14]. Similar enrichment strategies involving the SUMO3 T90K mutant and the anti-diglycine antibody [15], or a His10-Lys-deficient SUMO mutant with Ni-NTA enrichment [16] have also enabled the identification of thousands of SUMO sites. More recently, a multi-step approach combining free lysine acetylation followed by SENP2-cleavage of the fully acetylated SUMO2 enabled the identification of 751 putative wild-type SUMO-2 conjugation sites [17].
The presence of a side chain on SUMOylated peptides confer unusual structural features compared to other types of modifications such as acetylation, methylation, or phosphorylation. Indeed, the corresponding branched peptides typically expose a side chain amino group that provides an additional protonation site, and SUMOylated peptides have a propensity to form abundant triply protonated ions in LC-MS/MS experiments [18]. Furthermore, the presence of the side chain sequence also contributes to changes in the ion mobility of the peptide ion compared to its linear counterpart. These structural features were advantageously exploited in traveling wave ion mobility mass spectrometry to separate SUMO peptides from the typically smaller and lesser charged linear peptides [19].
Other forms of ion mobility such as high field asymmetric waveform ion mobility spectrometry (FAIMS) can be used to facilitate the identification of SUMOylated peptides in large-scale SUMO proteome analyses. In the context of proteomic analyses, FAIMS provides additional advantages as it can separate and accumulate multiply charged ions from co-eluting ions and background contaminants thereby improving MS sensitivity [20,19,22]. In FAIMS, ions are entrained by a carrier gas between two electrodes to which is applied a high voltage asymmetric waveform. Ions are separated in the gas phase based on their difference in mobility at low and high electric fields [23]. Ion selection is achieved by applying a compensation voltage (CV) that is superimposed to the waveform and enables the transmission of specific ions at their corresponding CV values. Previous reports have successfully combined FAIMS in LC-MS/MS analyses to expand the depth and comprehensiveness of proteomic analyses [24,23,24,27]. FAIMS also provides additional advantages in proteomics by separating phosphopeptides and peptide isomers [28,27,30], and by reducing the extent of precursor co-fragmentation to improve quantitative measurements of multiplex proteomics [31].
In the present study, we examined the mobility characteristics of SUMOylated peptides separated by FAIMS, and evaluated the analytic merits of this gas phase fractionation method when combined to LC-MS/MS. We further demonstrate the application of LC-FAIMS-MS/MS to expand the coverage and the dynamic range of the SUMO proteome of HEK293 cells exposed to heat shock.
Experimental
Cell Culture and Heat Shock
HEK293 wild-type cells and HEK293 cell line stably expressing 6xHis-SUMO-3-Q87R-Q88N (SUMO3m) [11] were cultured at 37 °C in a 5% CO2 constant atmosphere. Dulbecco’s Modified Eagles Medium (high glucose, GE Healthcare HyClone) was supplemented with 10% fetal bovine serum (Seradigm VWR Life Science), 1% Penicillin/Streptomycin Solution (Gibcon), and 1% L-Glutamine (Gibcon). For HEK293 SUMO3m, 0.5 mg ml−1 Neomycine (Gibcon) was added during cell culture. Cells were collected and washed twice with PBS (GE Healthcare HyClone) before mechanical lyses (2 × 10 s sonicate pulses) in 50 mM Tris in 8 M Urea (Bio Basic). Protein concentration was determined by Bradford assay (Bio Rad).
For heat stress experiments, HEK293 SUMO3m cells were grown in 15-cm petri plates to near confluency (0.7–0.8); the media was removed and replaced by media prewarmed at 43 °C without Neomycine. Cell dishes were placed in incubators for 60 min at 43 °C and 5% CO2. Control and heat shock-treated cells were collected simultaneously and washed twice with 37 °C PBS. Cell pellets were frozen in liquid nitrogen and stored at − 30 °C until further processing.
Synthetic Peptides
SUMOylated synthetic peptides were obtained from JPT Peptide Technologies GmbH (Berlin, Germany). Synthesis details were reported previous by our group [18]. For linearity experiment, each synthetic peptide was spiked at concentrations of 0.02 to 1 pmol in 760 ng of HEK293 protein digests. For HEK293 digests, 1 mg of proteins was resuspended in 1 mL of 50 mM ammonium bicarbonate (Sigma-Aldrich) and reduced for 30 min at 37 °C with 5 mM Tris (2-Carboxyethyl) phosphine Hydrochloride (TCEP) (Thermo Fisher Scientific). Alkylation reactions were performed for 30 min at room temperature with 10 mM 2-chloroacetamide (Sigma-Aldrich). Proteins were digested overnight at 37 °C with trypsin at a 1:50 trypsin:protein (wt/wt) (Promega, Madison, WI). Samples were dried down in a speed vacuum prior to their reconstitution in 0.2% formic acid at a final concentration of 1 mg/mL.
SUMO Peptide Enrichment
SUMOylated peptides from control or heat shock-treated cells were enriched according to the protocol described previously [15]. Briefly, cell pellets were lysed in 5 ml buffer A (6 M Guanine, 0.1 M NaH2PO4, 0.01 M Tris-HCl pH 8, 0.01 M Imidazole, 0.02 M 2-chloroacetamide, 0.01 M β-mercaptoethanol) followed by three short sonication pulses of 5 s to shear the DNA. Protein concentrations were determined by Bradford assay. SUMOylated proteins were purified by nickel affinity chromatography through the 6xHis tag on SUMO3m. 70 mg of protein from each condition (control or heath shock-treated cells) were immobilized on Ni-NTA Agarose (Qiagen) beads. For each sample, 1.5 mL of equilibrated Ni-NTA agarose beads (3 mL of Ni-NTA slurry that was previously washed four time with 10 mL buffer A) were incubated overnight with 70 mg of total cell lysate at 4 °C. The SUMO bound beads were washed once with 10 mL buffer A, five times with 10 mL buffer B (8 M Urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl pH 6.3, 0.01 M Imidazole, 0.01 M β-mercaptoethanol), twice with 50 mM Ammonium bicarbonate and finally resuspended in 3.5-mL 50 mM ammonium bicarbonate for quantification by Bradford assay. Proteins were digested on beads overnight at 37 °C with trypsin (Promega) at a 1:50 trypsin:protein (wt/wt) ratio. Peptides were acidified by adding TFA (Sigma-Aldrich) to final concentration of 1% and desalted on Oasis HLB columns (Waters). Peptides were dried down using a speed vacuum.
SUMOylated peptides were immuno-purified (IP) from the Ni-NTA enriched material using the anti-K(NQTGG) monoclonal antibody that was crosslinked to protein A magnetic beads [32]. Briefly, the peptides were resuspended in 1 mL of PBS and incubated with the anti-K(NQTGG) bound beads for 1 h at 4 °C at a 1:200 antibody:protein starting material (wt:wt). The immuno-complexes were washed three times with 1 mL of PBS, twice with 1 mL of 0.1 × PBS and once with 1 mL of water. The SUMOylated peptides were eluted from the beads with 500-μL 0.2% formic acid and dried down in a speed vacuum. The peptides were reconstituted in 4% formic acid in water for MS analysis (6 mg of starting material per injection).
Immunoblot Analysis
Total cell extracts (10 μg) were prepared in Laemmli buffer (0.06 M Tris-Cl pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue) and resolved on a precast gel (4–12% Bis-Tris) (Criterion XT). Proteins were transferred from the gel onto a nitrocellulose membrane at 100 mA for 16 h. The membrane was blocked with 5% non-fat milk and incubated with the primary antibody (SUMO2/3 or Histone H3, cell Signaling). Bands were visualized using the ECL chemiluminescence detection methodology while using an anti-rabbit horseradish peroxidase-conjugated secondary antibody.
Mass Spectrometry
LC-MS/MS analyses were performed on a nano-LC 2D pump (Eksigent, Dublin, CA) interfaced to a LTQ-Orbitrap Elite hybrid mass spectrometer (Thermo Fisher Science, San Jose, CA). On-line peptide separation was performed on an Optiguard SCX trap column, 5 μm, 300 Å, 0.5 ID × 23 mm (Optimize Technologies, Oregon City, OR) followed by a 360-μm ID × 4 mm, C18 trap column before separation on an in-house packed 150 μm ID × 20 cm LC column (Jupiter C18, 3 μm, 300 Å, Phenomenex, Torrance, CA). For two-dimensional chromatography, peptides were loaded with 0.2% formic acid to the SCX cartridge and eluted using increasing concentration of ammonium acetate (250, 500, 750, 1000, and 2000 mM) at pH 3.5. Reversed phase separation was performed with a linear gradient of 5–40% acetonitrile (0.2% formic acid) at a flow rate of 600 nL/min. For synthetic peptides, HEK293 and HEK293 SUMO3m digests, the separation was achieved with 56- or 106-min long gradient, respectively, before re-equilibrating the column for 14 min at 5% acetonitrile (0.2% formic acid). MS/MS scans were triggered using a top 12 method; MS scans were acquired in the orbitrap at a resolution of 60,000 whereas the resolution for MS/MS spectra was at 15000. Full MS were scanned between 300 and 1200 m/z and the normalized collision energy for high-energy collision-induced dissociation fragmentation was set at 30. Automatic Gain Control for full MS and MS/MS were set at 106 and 5 × 104, respectively with exclusion times of 45 s (without FAIMS) or 20 s (with FAIMS). The maximum injection time for the full MS was 1 s, whereas for MS/MS the maximum injection times were set to 300 ms.
FAIMS
For LC-FAIMS-MS/MS measurements, the Ion Max Source (Thermo Fisher Science, San Jose, CA) was replaced with a FAIMS interface (Thermo Fisher Science, San Jose, CA). Experiments were conducted on a FAIMS device with a curved well ion inlet [33] with two cylindrical electrodes with a gap width of 1.5 mm between inner and outer electrodes at 90 and 100 °C, respectively. Dispersion voltage (DV) was held at − 5000 V (ED − 3333 V/cm). The FAIMS interface used nitrogen as a carrier gas at a flow rate of 2.3 L/min. For electrospray ionization, a cylindrical stainless steel tube was connected to the analytical column outlet and mounted at a 60° angle. MS parameters for LC-FAIMS-MS/MS measurements were similar to those described above, except that the top 12 method (1 MS survey scan followed by a maximum of 12 tandem MS2) was replaced with three CV step scan method (one MS survey scan followed by three tandem MS2 for each CV step). For synthetic peptides, the CV range from − 34 to − 85 V (− 227 to − 567 V/cm) was covered, for large-scale experiment where cells were exposed to heat stress, four injections covered the range from CV − 40 V to CV − 73 V (− 267 to − 487 V/cm) using 3 V (20 V/cm) increments.
Data Processing
Raw data acquired with Xcalibur software (version 2.2) were either processed with MaxQuant version 1.5.3.8 (http://www.maxquant.org) or Mascot Daemon (Matrix Science, version 2.4.0). SUMOylated peptides isolated by immunoprecipitations were searched using MaxQuant and the Uniprot Human database (release March 3, 2015). FAIMS Raw files were transformed with an inHouse python script to mzXML files containing only one CV per file. Search parameters were as follows: Trypsin/P as specific enzyme, Oxidation (M)–Phospho (STY)–Deamidation (NQ)–GlyGly (K) and SUMO3: NQTGG (K) as variable modification. The tolerance for MS and MS/MS were set to 25 and 20 ppm for the first search and 6 and 10 ppm for the main search. MS/MS identification was filtered with an FDR of 0.1%. For network analysis, gene names were searched against the whole human proteome using string version 10.5 (https://string-db.org). Experimentally determined interactors with high confidence (0.700) were visualized in Cytoscape (Version 3.5.1) and clustered with the MCODE application (degree Cutoff 2, Node Score Cutoff 0.1, K-Core 2 and Max.Depth 10). Gene ontology enrichment analysis of the extracted clusters was performed with BiNGO with a significance level of 0.05.
Results and Discussion
The strategy to identify SUMOylated proteins and their modified sites relies on HEK293 cells stably expressing a functional SUMO3 mutant that contains an N-terminal 6xHis tag and a C-terminus cleavage site that leaves a five amino acid SUMO remnant (NQTGG) on the lysine acceptor site upon tryptic digestion [10, 11]. While this protocol provides enrichment level up to 50%, the identification of SUMOylated peptides in a progressively more complex population of low abundance peptides still represent a sizable analytical problem. In this context, we surmised that the ability of FAIMS to separate ions based on their charge states and their differences in mobility at low and high electric fields may provide a unique advantage to facilitate the detection of low abundance SUMOylated peptides. We reasoned that the branched structure of SUMOylated peptides and the presence of an extra N-terminus group confer distinctive features that favors their separation from linear tryptic peptides (Figure 1a). Consequently, we evaluated the distribution of SUMOylated peptides across CV values to determine conditions under which target peptide ions can be transmitted for enhanced SUMO proteome coverage.
Gas-phase separation of SUMOylated peptides using FAIMS. (a) Schematic representation of regular tryptic peptides and peptides modified by SUMO3 with remnant NQTGG. (b) Infusion of five SUMO peptides separated between two FAIMS electrodes with a gap of 1.5 mm. (c) Intensity and abundance distribution for the 5 SUMO peptides. Conditions: N2 carrier gas 2.3 L/min, DV − 5000 V
FAIMS Improves the Detection and Identification of SUMOylated Peptides Present in Complex Tryptic Digests
In preliminary experiments, we infused a mixture of five synthetic SUMOylated peptides at 100 nM with and without FAIMS. To increase both ion transmission and resolution, we used FAIMS electrodes with a narrow gap of 1.5 mm that provides higher resolution and lower gas flow turbulences compared to previous generation devices [33]. This FAIMS device provides a larger range of CV distribution extending over more than 60 V and uses nitrogen only as carrier gas [34]. A plot of the signal intensities versus CV values is shown in Figure 1b, and highlights the ability of FAIMS to separate triply charged SUMOylated peptides from their doubly charged counterparts. As indicated, the triply charged ions are transmitted at higher CV values between − 40 and − 80 V whereas doubly charged ions were observed between − 16 and − 32 V. Also, the triply charged peptide ions were resolved from each other compared to doubly charged ions that were all transmitted at similar CVs with limited separation except for peptide LLVHMGLLKSEDK (italic type indicated the modified residue). A comparison of the intensity of the SUMOylated peptides analyzed with and without FAIMS revealed that higher transmission was generally obtained when using gas phase ion fractionation with FAIMS (Figure 1c). This observation might reflect the selective enrichment of target ions for a fix ion trapping capacity when using FAIMS. Interestingly, we noted that the relative proportion of doubly and triply charged peptide ions varied between experiments, and that the latter ions showed a higher transmission when using FAIMS. The transmission bias of doubly charged SUMOylated peptides observed without FAIMS might be explained by suppression of higher charge states that could take place when approaching the ion capacity defined by the automatic gain control (AGC).
To investigate the impact of sample complexity on the identification of multiply charged SUMOylated peptide ions, we infused the synthetic SUMO peptide LLVHMGLLKSEDK at a constant concentration of 2 μM with increasing amounts of HEK293 tryptic digest (Supplementary Figure 1). Without any HEK293 digest, the quadruply charged peptide ion was the most abundant charge state (57% relative abundance compared with 2+ ion at 5%). However, the relative proportion of this ion was progressively reduced with increasing contribution of HEK293 digest, and represented only 31% when spiked in 4 μg of HEK293 digest, whereas its doubly charged ion increased to 33%. The discrimination of higher charge states with increasing sample complexity was also observed at different AGC values, but was more significant at AGC 104 compared to an AGC of 107 (Supplementary Figure 2). These results confirm that the distribution and intensity of multiply charged SUMO peptide ions is affected by increasing sample complexity, a situation that may impede their successful identification when present at low abundance in cell digests.
To determine if suppression effects undermine the detection of SUMOylated peptides, we performed LC-MS/MS experiments with and without FAIMS on a mixture of 154 synthetic SUMO peptides and a tryptic digest of HEK293 cells. To ensure that we maximize the number of identification, we performed triplicate LC-MS/MS experiments for synthetic SUMO peptides and 12 replicates for the HEK293 digest. The same samples were analyzed by LC-FAIMS-MS/MS except that the number of CV acquired was scaled according to sample complexity (i.e., injection at each CV for HEK293 digest, and every 3 V increments for synthetic SUMO peptides). For the HEK293 digest, LC-MS/MS performed with and without FAIMS enabled the identification of 14,442 and 6103 unique peptides, with an overlap of 4499 peptides. Out of these common peptides, only 1173 and 993 peptides were identified as doubly and triply charged peptide ions in FAIMS and non FAIMS experiments, respectively. The LC-MS/MS analyses of synthetic SUMOylated peptides with and without FAIMS identified 118 and 144 peptides, of which 108 were common. However, only 15 and 25 of these common peptides displayed both doubly and triply charged ions with and without FAIMS, respectively. Next, we determined the intensity ratio of doubly and triply charged ions for all identified peptides. The boxplot distribution of the corresponding ratios indicates that HEK293 tryptic peptides analyzed without FAIMS show an almost equal intensity of doubly and triply charged ions whereas the latter ions were more abundant when using FAIMS (Figure 2a). This intensity bias with and without FAIMS was also observed for SUMOylated peptides where the intensity of triply charged ions was more intense than their doubly charged counterparts, and suggest that the underrepresentation of multiply charged ions is correlated with increasing sample complexity.
Charge state distribution of tryptic peptides and SUMOylated peptides. (a) Distribution of intensity ratios between doubly and triply protonated peptide ions from a HEK293 tryptic digest and SUMOylated peptides. (b) Bar chart representation of charge states vs CV values for tryptic peptides from HEK293 cells and SUMOylated peptides. (c) Density plot showing the distribution of m/z vs. CV for tryptic peptides from HEK293 cells and SUMOylated peptides
The ability of FAIMS to separate ion populations into different CV regions enables the selective transmission of triply charged ions with limited contribution of lower charge state ions. This is illustrated in Figure 2b that compares the distribution of multiply charged ions identified with FAIMS according to their CV values. As indicated, a larger proportion of triply charged peptides, including SUMOylated peptides, are transmitted between − 37 and − 70 V, in a region where reduced contribution of doubly charged ions is typically observed. While triply charged peptides ions from HEK293 and synthetic SUMOylated peptides are transmitted across a similar range of CV, the branched structure and the charge distribution of the latter peptides confer notable differences in mobility that can be advantageously exploited to facilitate their separation from the bulk of tryptic peptides. Figure 2c shows the density plots of m/z distribution vs. CV values for HEK293 and synthetic SUMOylated peptides. As anticipated, SUMOylated peptides are clearly distinct from the larger population of tryptic peptides based on their distribution of m/z and CV values. A more detailed examination of the data also revealed that 3+ ions from SUMO peptides were transmitted at lower CV values than 3+ tryptic peptides over the m/z range examined. However, the difference in mean CV values at which SUMO and tryptic peptide 3+ ions were transmitted gradually decreased with increasing m/z values (Supplementary Figure 3). These observations suggest that conformational changes imparted by branched SUMO peptides have more important effects on changes in ion mobility at high and low electric field than tryptic peptide ions of similar masses. Therefore, the selection of appropriate CV values could favor the transmission of SUMOylated peptides to enhance their identification in large-scale SUMO proteome analyses.
We next evaluated the linearity and limits of detection (LOD) for the analysis of a mixture of SUMOylated peptides spiked at levels of 20 fmol to 2 pmol in a tryptic digest of HEK293 proteins (760 ng) using LC-MS/MS with and without FAIMS (Figure 3a). Triplicate measurements were obtained for all experiments. For LC-FAIMS/MS/MS, three CV values were selected per injection to cover the range of − 34 to − 80 V in 3 V increments. In total, we identified 107 synthetic SUMOylated peptides common to each experiment corresponding to 69.5% of the total number of spiked peptides, and the distribution of the linear regression coefficients of determination (r2) are shown in Figure 3b. Similar distribution of r2 values were obtained for LC-MS/MS experiments performed with and without FAIMS. Examination of all linear regression analyses indicates that the CV stepping using in LC-FAIMS-MS/MS has no substantial effect on the linear regression (Supplementary Figure 2). However, analyses performed using FAIMS provided significant gains in sensitivity that were reflected with lower LOD values. An example of this is shown in Figure 3c for the triply protonated peptide ion at m/z 576.65 corresponding to the synthetic SUMO peptide INEILSNALKR spiked at a level of 200 fmol. It is noteworthy that the LC-MS/MS analysis performed without FAIMS showed the co-elution of another peptide ion at m/z 577.32 which gave rise to a chimeric tandem mass spectrum, and prevented the non-ambiguous identification of the corresponding SUMOylated peptide. In contrast, the same analysis performed using FAIMS showed a distinct signal at m/z 576.65 with a clear improvement in signal to noise that translated into an ~ 10-fold improvement in LOD. A comparison of the LOD values obtained for all detected SUMOylated peptides is shown in Figure 3d. Of the 107 SUMOylated peptides quantified using LC-MS/MS, 100 showed an improvement of LOD that ranged from 2- to 228-fold when using FAIMS. Marked improvements of sensitivity were noted for SUMO peptides of lower molecular mass that are observed in more densely populated region of the mass spectrum where co-elution of abundant peptide ions with similar m/z values can affect peak detectability (Figure 3c).
LC-MS/MS analyses of SUMOylated peptides spiked in a HEK293 tryptic digest. (a) Synthetic SUMOylated peptides were spiked at concentration ranging from 0.02 to 1.00 pmol in 760 ng of a HEK293 tryptic digest. (b) Distribution of r2 values for all identified SUMOylated peptides analyzed by LC-MS/MS with and without FAIMS. (c) Narrow section of the mass spectrum showing m/z 576.650 3+ corresponding to the peptide INEILSNAKR with and without FAIMS. (d) Scatter plot comparing the limit of detection of SUMOylated peptides detected in both LC-MS/MS experiments
Large-Scale Profiling of SUMO Proteome Under Heat Shock
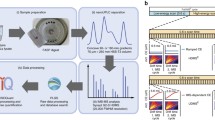
To evaluate the analytical merits of FAIMS for the identification of SUMOylated proteins in a site-specific manner, we profiled the changes in protein SUMOylation of HEK293 SUMO3m cells upon heat shock. Protein SUMOylation is an important modification regulating the cellular defense against hyperthermic cytotoxicity. For example, several transcription factors are known to be SUMOylated in response to heat shock, a modification that impedes their functions via several mechanisms including reduced nuclear entry, inhibition of DNA binding, and recruitment of transcriptional repressor [35]. Global changes in protein SUMOylation is rapidly observed upon heat shock treatment [36]. Accordingly, we performed label-free quantitative proteomic analyses on HEK293 SUMO3m cells subjected to 43 °C heat stress for 1 h (Figure 4a). Increase in global protein SUMOylation upon heat shock was monitored by western blot, as depicted in Figure 4b. We selected this incubation period based on the progressive increase in protein SUMOylation that reached a plateau after 1 h (Supplementary Figure 5). Control (CTL) and heat shock-treated HEK293 SUMO3m cells were harvested and lysed in denaturing buffer. SUMOylated proteins were then enriched on a Ni-NTA column prior to trypsin digestion (Figure 4c). SUMOylated tryptic peptides were purified by immunoaffinity using a custom antibody that recognizes the five amino acid SUMO3m remnant epitope [15]. The CTL and heat shock samples were then fractionated by on-line SCX and analyzed by LC-MS/MS or LC-FAIMS-MS/MS on the Orbitrap Elite mass spectrometer (Figure 4c). A total of six replicate injections were conducted on each SCX fractions analyzed by LC-MS/MS, while a single injection at different CV values was performed for individual SCX fraction analyzed by LC-FAIMS-MS/MS.
SUMO proteome analysis of HEK293 cells following heat shock. (a) HEK293 cells overexpressing SUMO3m were cultured at 37 °C. Control cells were kept at 37 °C. For heat shock experiments the culture media was removed and replaced with preheated media and incubated for 1 h at 43 °C. (b) Western blot of control and heat shock-treated HEK293 cells using a SUMO2/3 specific antibody. The western blot of histone H3 is shown as a loading control. (c) Overview of the immunoaffinity purification and LC-MS/MS analyses of SUMOylated peptides. Total cell lysates are subjected to a NiNTA column to enrich SUMOylated proteins before tryptic digestion. Peptides containing the SUMO3m remnant are enriched using the anti-K-(NQTGG) antibody prior to their analyses using on-line SCX fractionation and LC-MS/MS with and without FAIMS
The large-scale SUMO proteome analyses of HEK293 SUMO3m cells identified a total of 3658 unique SUMO sites on 1433 protein substrates (Figure 5a, Supplementary table I). FAIMS enabled a 36% increase in SUMO proteome coverage compared to the conventional LC-MS/MS approach with 2820 SUMO sites (1254 SUMOylated proteins) and 2078 SUMO sites (951 SUMOylated proteins) for FAIMS and non FAIMS experiments, respectively. These experiments also highlighted that the number of SUMO sites identified by both approaches represent only ~ 1/3 of all identified SUMO sites. In an effort to understand the reasons behind these differences, we compared the distribution of charge states for SUMOylated peptides identified in FAIMS and non FAIMS experiments. Supplementary Figure 6a indicates that SUMOylated peptides uniquely identified in either FAIMS or non FAIMS experiments were predominantly triply charged ions. To determine if SUMOylated peptides unique to non FAIMS experiments were filtered out using FAIMS due to the CV steps selected, we used the “match between runs” function of Maxquant. This function enables the time and m/z alignment of peptide ions and the correlation of their intensities in a pairwise fashion for peptides identified in at least one of the two experiments. The correlation of detected peptides using “match between runs” indicated that 79.5% of all SUMOylated peptides were present in both FAIMS and non FAIMS experiments (Figure 5b). These analyses also revealed that unique peptides originally identified without FAIMS were detected in FAIMS experiments but that the converse was not necessarily true (Supplementary Figure 7). Indeed, SUMOylated peptides identified without FAIMS were detected in FAIMS experiments as a distinct isotopic cluster at an intensity level approaching the threshold for triggering MS/MS acquisition. However, SUMOylated peptides uniquely identified with FAIMS showed either interfering ions or were of too low intensity to be detected in non FAIMS experiments. Interestingly, we only observed a marginal increase in the number of SUMOylated peptides identified beyond three replicate injection of the SCX fraction without FAIMS, whereas a progressive gain in new identification was observed using FAIMS across different CV steps (Figure 5b and Supplementary Figure 6b).
SUMO proteome analyses of HEK293 cells following heat shock. (a) Venn diagram showing the distribution of SUMOylation sites and SUMOylated proteins identified with and without FAIMS. (b) Sum of unique SUMOylation sites identified after each replicate. (c) Enrichment of SUMOylated peptides across different ranges of CV values. (d) Distribution of unique SUMOylation sites as a function of CV values
The increased number of identified SUMOylated peptides using FAIMS is reminiscent of observations made during the large-scale analysis of tryptic peptides [37], where sample complexity limits the progressive increase of new identification for replicate injections performed without FAIMS. In this context, gas phase fractionation using FAIMS provides sufficient resolution to prevent the resampling of peptides across CV steps. Importantly, FAIMS enables the enrichment of SUMOylated peptides by favoring their selective transmission in a CV range that minimizes the contribution of interfering peptides ions (Supplementary Figure 6e).
Next, we compared the enrichment levels of SUMO peptides for FAIMS and non FAIMS experiments (Figure 5c). These analyses indicated that the proportion of SUMOylated peptides in non FAIMS experiments represented only 18% of all peptides identified. In contrast, FAIMS provided enrichment levels up to 68% as observed for high CV values (− 84, − 87, and − 90 V). Interestingly, the increase in enrichment level correlates well with the progressive increase in the number of unique SUMOylated peptides observed with FAIMS for higher CVs (Figure 5d).
The gas phase fractionation capability of FAIMS reduced sample complexity and enabled more comprehensive SUMO proteome analysis. To evaluate the extent of sample complexity, we determined the precursor intensity fraction (PIF) for all precursor ions acquired in FAIMS and non FAIMS experiments (Supplementary Figure 6c). The PIF represents the proportion of ion current in the isolation window associated with the target ion and is available in Maxquant [38]. A PIF value approaching 1 indicates that most of the ion current comes from the precursor of interest and contains a low proportion of co-isolated ions. When comparing the distribution of PIF values, we noted that FAIMS reduced significantly the proportion of contaminating precursor ions which decreased the occurrence of co-fragmentation and provided higher Andromeda scores (Supplementary Figures 6c, 6d). The intensity distribution of peptides identified only by FAIMS (Supplementary Figure 6f) highlights the ability of FAIMS to identify lower intensity peptides by reducing background level, consistent with that reported previously in large-scale proteomic experiments [37]. We noted that the intensity of the SUMO peptides common to both FAIMS and non FAIMS was 30% lower in the former experiment (Supplementary Figure 6g), suggesting that the distribution of selected CV steps and slower duty cycle resulted in reduced ion transmission.
To profile the changes in protein SUMOylation upon heat shock, we used a label-free quantitative proteomic approach and compared the extent of quantifiable peptides/proteins by LC-MS/MS with and without FAIMS. We quantified 2889 unique SUMO sites on 1235 protein substrates, of which 887 were common to both FAIMS and non FAIMS experiments. A comparison of the fold changes observed for the corresponding SUMOylated peptides indicated that the majority of quantified peptides displayed comparable changes in abundance between experiments (Figure 6a). These analyses revealed a global increase in protein SUMOylation upon 1 h heat shock treatment, consistent with the immunoblots shown in Figure 4b. However, peptides quantified using FAIMS displayed higher PIF values (median 0.95) compared to those from non FAIMS LC-MS/MS experiments (median 0.84). An example is shown in Figure 6b for the quantification of the SUMOylated peptide FFSCDKIQNGAQGIR from the heterogeneous nuclear ribonucleoprotein H1. This peptide was identified in FAIMS and non FAIMS experiments with similar changes in abundance upon heat shock (~ 15-fold), though the lower PIF values observed in conventional LC-MS/MS resulted in a mixed MS/MS spectrum with a lower identification score (Figure 6b) .
Profiling changes in protein SUMOylation of HEK293 cells following heat shock. (a) Comparison of fold change measurements for common SUMOylated peptides identified with and without FAIMS. (b) MS/MS spectra for FFSCDKIQNGAQGIR and the corresponding extracted ion chromatogram for LC-MS/MS analyses with (bottom) and without FAIMS (top). (c) Distribution of fold change and SUMOylation sites rank plot showing sites for transcription factors in red. (d) Number of SUMOylated sites per target proteins. Poly [ADP-ribose] polymerase 1 (PARP-1) shows the highest number of SUMOylation with 40 different sites
Overall, we observed that approximately two third of the SUMOylation sites were up-regulated in response to the heat shock with both approaches (Figure 6c). Heat shock typically results in an accumulation of misfolded proteins and elevated levels of protein aggregates. To maintain their viability during the heat stress, cells inhibit the transcription, and translation machinery whereby several transcription factors (TFs) are SUMOylated [39,38,41]. TFs are known to be an important target of SUMOylation, predominantly resulting in suppression of transcription and regulation of binding of TFs to chromatin [42]. In contrast, heat shock factors 1 and 2 (HSF1 and HSF2) which play a central role in the transcriptional activation of the heat shock response [39, 43] are activated, and promote the expression of heat shock proteins that act as molecular chaperone to facilitate the refolding of damaged/misfolded proteins [44]. Our SUMO proteome analyses revealed that SUMOylation on K126 and K131 of HSF1 and K2, K82, K139, and K151 of HSF2 showed more than two-fold increase in SUMOylation upon heat shock. Likewise, most TFs showed a high increase in SUMOylation following heat shock (Figure 6c). A specific example is the multifunctional general transcription factor II-I (GTF2I), a protein known to play a role in cell cycle regulation and the DNA damage response pathway. In total, 34 SUMO sites were identified on this protein, half of which were observed only after heat stress. Although the increase in the SUMOylation of GTF2I has been observed in response to various stressors, the function of its SUMOylation is still elusive [45].
Cells can also survive the heat stress by other means, such as promoting the degradation of misfolded proteins via the ubiquitin-proteasome machinery. Earlier reports indicated that promyelocytic leukemia protein (PML) recognizes misfolded nuclear proteins and initiates their SUMO2/3 conjugation [46]. This suggests that SUMOylation of specific proteins by SUMO2/3 is essential for their subsequent conjugation to ubiquitin via the ubiquitin E3 activity of the SUMO-targeted ubiquitin ligase RNF4, leading to the proteosomal degradation of their SUMOylated targets [47]. Our high confidence dataset (0.1% FDR) contains 3716 unique SUMO sites from 1564 target proteins, 46% with multiple SUMO sites (≥ 2 SUMO 3 sites per protein). For example, multi-SUMOylation was detected on proteins nvolved in the SUMOylation pathway (SUMO1, SUMO2, SUMO3, PIAS1, PIAS4, UBA2, PML, TRIM28, SMARCA4, SMC5, SMC6, NSMCE2, BRCA1, BLM WRN, PARP1, and RANBP2), where TOPORS, PIAS1, PIAS4, and NSMCE2 are SUMO E3 ligases. Among all SUMO targets, 263 contained four or more SUMO sites, including poly(ADP-ribose) polymerase (PARP1), a protein known to be SUMOylated [48] (Figure 6d). PARP1 was previously known to be multi-SUMOylated, and our experiments identified 40 SUMOylation sites on PARP1, the majority of which were strongly up-regulated after heat shock. More than 12% of the amino acid sequence of PARP1 is composed of lysine residues and harbor more than 20 putative SUMO consensus motifs, including K203 and K249 residues located within its DNA-binding domain, K486 and K512 are found in its auto-modification domain and K798 is in its catalytic domain [48]. PARP1, which is the most abundant member of the PARP family [49], plays an important role in DNA repair where it recognizes DNA damage and initiates the repair response for single- or double-strand breaks [50]. Furthermore, PARP1 is modified by several PTM such as phosphorylation, acetylation, and ubiquitination in addition to its SUMOylation [49]. In our dataset, 21 out of the 40 SUMOylation sites identified on PARP1 showed more than 2-fold change in abundance upon heat stress. Martin et al. reported that multi or polySUMOylation of PARP1 was initiated by PIAS4 [48]. Moreover, the SUMO targeted ubiquitin ligase RNF4 recognizes SUMOylated PARP1 and mediates its proteasome degradation. This cascade promotes the heat shock response through the activation of the HSP70.1 promoter [48].
The SUMOylated proteins were subjected to a network analysis. Clusters of tightly interacting proteins were extracted from this network for gene ontology analysis [11, 17]. Ribosomes that are involved in translation elongation form the largest cluster group, together with proteins involved in ribosome biogenesis. FAIMS extended the coverage of the ribosome biogenesis cluster and identified nucleolus proteins DCAF13, WDR36, WDR46, and NOL6 (Figure 7). Moreover, a deeper coverage of the proteasome complex was achieved with FAIMS (PSMD8, PSMA4, PSMA5, PSMA7, SHFM1, UBLCP1, and BAP1 were identified with FAIMS whereas PSMD11 and PSMD12 were unique to non FAIMS). Interestingly, our group showed that the SUMOylation of the proteasome is greatly induced during an MG132 treatment, which leads to the discovery that the SUMOylated proteasome is shuttled to the PML nuclear bodies through its SUMO interacting motif, thereby promoting the degradation of SUMOylated proteins found in the PML nuclear bodies [15]. This hearkens back to the notion that cells can overcome heat stress by degrading misfolded proteins, in this case in a SUMO dependent manner. Moreover, BAP1, which was found only in the FAIMS dataset, encodes the ubiquitin carboxy-terminal hydrolase BRCA1 associated protein-1. BRCA1 is a SUMO-regulated ubiquitin ligase (SRUbL), where SUMOylation of BRCA1 promotes its ubiquitinylation activity [51]. BRCA1 is a co-factor of GTF2I [45] and is involved in the DNA damage response (Supplementary Figure 8). Interestingly, the SUMOylation of several proteins involved in the DNA damage response pathway were upregulated during heat shock, and is consistent with previous reports indicating that hyperthermia induces DNA double-strand breaks (DSBs) in mammalian cells [52, 53].
Proteins involved in chromatin modification/organization through the SWI/SNF chromatin remodeling complex also showed an increase in SUMOylation upon heat shock. Several modification sites were located near or within DNA-binding domain and could affect the transcriptional activities. For example, we noted that upon heat shock the transcriptional activator BRG1 (SMARCA4) was SUMOylated at residue K711 proximal to the helicase ATP-binding domain and at K1131 within the helicase C-terminal domain. The SUMOylation of SMARCA4 at these residues could impede its helicase activity. Similarly, we identified the increase in SUMOylation of several members of the NuRD complex that control chromatin remodeling through the use of histone deacetylases (HDACs). For example, HDAC1 was SUMOylated at ten residues including K66, K412, K467, and K476 that showed more than 4-fold up-regulation. Previous studies reported that the SUMOylation of HDAC1 is required to repress transcriptional activities at defined promoters [54, 55]. However, it is still unclear if SUMOylation directly impedes HDAC activity or mediate the recruitment of other chromatin repressors.
Conclusion
This study shows that gas phase ion fractionation using FAIMS can provide a unique advantage in SUMO proteome analyses by favoring the transmission of branched SUMOylated peptides that can be underrepresented in complex tryptic digests even after immunoaffinity enrichment. When compared to tryptic peptides, SUMOylated peptides show a higher propensity to form triply charged ions that can be selectively transmitted by FAIMS at CV values where fewer linear tryptic peptides are present. The reduced sample complexity observed at the corresponding CV values confers two important advantages in proteomics. First, it improves the detection and identification of low abundance peptides such as those modified by SUMOylation as shown in the present study. Gas phase ion fractionation using FAIMS also facilitates the detection of multiply charged ions that could be suppressed when present in a complex population of co-eluting peptides. The reduced contribution of co-eluting ions and contaminants observed when using FAIMS significantly improved the quality of the MS/MS spectra and led to a 36% improvement in SUMO proteome coverage compared to non FAIMS experiments. Secondly, the reduced sample complexity also enabled the quantification of a larger number of SUMOylated peptides and provided more accurate measurements of changes in protein SUMOylation. While the present study used label-free quantitative proteomics, marked improvements in precision of quantitative measurements were previously reported for multiplex isobaric labeling when using FAIMS. We anticipated that the availability of FAIMS on new generation of mass spectrometers that provide higher resolution and acquisition rates will significantly expand the depth and coverage of proteome analyses and will facilitate the profiling of low abundance protein modifications that remain inaccessible with current LC-MS/MS strategies.
References
Streich Jr., F.C., Lima, C.D.: Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 43, 357–379 (2014)
van der Veen, A.G., Ploegh, H.L.: Ubiquitin-like proteins. Annu. Rev. Biochem. 81, 323–357 (2012)
Dou, H., Huang, C., Van Nguyen, T., Lu, L.S., Yeh, E.T.: SUMOylation and de-SUMOylation in response to DNA damage. FEBS Lett. 585, 2891–2896 (2011)
Niskanen, E.A., Malinen, M., Sutinen, P., Toropainen, S., Paakinaho, V., Vihervaara, A., Joutsen, J., Kaikkonen, M.U., Sistonen, L., Palvimo, J.J.: Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol. 16, 153–172 (2015)
Tatham, M.H., Matic, I., Mann, M., Hay, R.T.: Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci. Signal. 4(1), (2011)
Eiffer, K., Vertegaal, A.C.O.: Mapping the sumoylated laadscape. FEBS J. 282, 3669–3680 (2015)
Hay, R.T.: Decoding the SUMO signal. Biochem. Soc. Trans. 41, 463–473 (2013)
Hay, R.T.: SUMO: a history of modification. Mol. Cell. 18, 1–12 (2005)
Tatham, M.H., Jaffray, E.G., Vaughan, O.A., desterro, J.M., Bottino, C.H., Naismith, J.H., Hay, R.T.: Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35374 (2001)
Hendricks, I.A., Lyon, D., Young, C., Jensen, L.J., Vertegaal, A.C., Nielsen, M.L.: Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 24, 325–336 (2017)
Hendricks, I.A., Vertegaal, A.C.: A comprehensive compilation of SUMO proteomics. Nat. Rev. 17, 581–595 (2016)
Hendriks, I.A., D’Souza, R.C., Yang, B., Verlaan-de Vries, M., Mann, M., Vertegaal, A.C.: Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 21, 927–936 (2014)
Galisson, F., Mahrouche, L., Courcelles, M., Bonneil, E., Meloche, S., Chelbi-Alix, M.K., Thibault, P.: A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol. Cell. Proteomics. 10, (2011)
Lamoliatte, F., Caron, D., Durette, C., Mahrouche, L., Maroui, M.A., Caron-Lizotte, O., Bonneil, E., Chelbi-Alix, M.K., Thibault, P.: Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat. Commun. 5, 5409–5420 (2014)
Lamoliatte, F., McManus, F.P., Maarifi, G., Chelbi-Alix, M.K., Thibault, P.: Uncovering the SUMOylation and Ubiquitylation crosstalk in Human Cells Using Sequential Peptide Immunopurification. Nat. Commun., (2017)
Tammsalu, T., Matic, I., Jaffray, E.G., Ibrahim, A.F., Tatham, M.H., Hay, R.T.: Proteome-wide identification of SUMO2 modification sites. Sci. Signal. 7, (2014)
Hendriks, I.A., D’Souza, R.C., Chang, J.-G., Mann, M., Vertegaal, A.C.O.: System-wide identification of wild-type SUMO-2 conjugation sites. Nat. Commun. 6, 7289–7305 (2015)
Lamoliatte, F., Bonneil, E., Durette, C., Caron-Lizotte, O., Wildemann, D., Zerweck, J., Wenshuck, H., Thibault, P.: Targeted identification of SUMOylation sites in human proteins using affinity enrichment and paralog-specific reporter ions. Mol. Cell. Proteomics. 12, 2536–2550 (2013)
Dumont, Q., Donaldson, D.L., Griffith, W.P.: Screening method for isopeptides from small ubiquitin-related modifier-conjugated proteins by ion mobility mass spectrometry. Anal. Chem. 83, 9638–9642 (2011)
Barnett, D.A., Ding, L., Ells, B., Purves, R.W., Guevremont, R.: Tandem mass spectra of tryptic peptides at signal-to-background ratios approaching unity using electrospray ionization high-field asymmetric waveform ion mobility spectrometry/hybrid quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 16, 676–680 (2002)
Barnett, D.A., Ells, B., Guevremont, R., Purves, R.W.: Application of ESI-FAIMS-MS to the analysis of tryptic peptides. J. Am. Soc. Mass Spectrom. 13, 1282–1291 (2002)
Purves, R.W., Guevremont, R.: Electrospray ionization high-field asymmetric waveform ion mobility spectrometry-mass spectrometry. Anal. Chem. 71, 2346–2357 (1999)
Guevremont, R.: High-field asymmetric waveform ion mobility spectrometry: a new tool for mass spectrometry. J. Chromatogr. A. 1058, 3–19 (2004)
Creese, A.J., Shimwell, N.J., Larkins, K.P.B., Heath, J.K., Cooper, H.J.: Probing the complementarity of FAIMS and strong cation exchange chromatography in shotgun proteomics. J. Am. Soc. Mass Spectrom. 24, 431–443 (2013)
Saba, J., Bonneil, E., Pomiès, C., Eng, K., Thibault, P.: Enhanced sensitivity in proteomics experiments using FAIMS coupled with a hybrid linear ion trap/orbitrap mass analyzer. J. Proteome Res. 8, 3355–3366 (2009)
Swearingen, K.E., Hoopmann, M.R., Johnson, R.S., Saleem, R.A., Aitchinson, J.D., Moritz, R.L.: Nanospray FAIMS fractionation provides significant increases in proteome coverage of unfractionated complex protein digests. Mol. Cellular Proteomics. (2012)
Xuan, Y., Creese, A.J., Horner, J.A., Cooper, H.J.: High-field asymmetric waveform ion mobility spectrometry (FAIMS) coupled with high-resolution electron transfer dissociation mass spectrometry for the analysis of isobaric phosphopeptides. Rapid Commun. Mass Spectrom. 23, 1963–1969 (2009)
Bridon, G., Bonneil, E., Muratore-Schroeder, T., Caron-Lizotte, O., Thibault, P.: Improvement of phosphoproteome analyses using FAIMS and decision tree fragmentation. Application to the insulin signaling pathway in Drosophila melanogaster S2 cells. J. Proteome Res. 11, 927–940 (2012)
Creese, A.J., Smart, J., Cooper, H.J.: Large-scale analysis of peptide sequence variants: the case for high-field asymmetric waveform ion mobility spectrometry. Anal. Chem. 85, 4836–4843 (2013)
Zhao, H., Cunningham, D.L., Creese, A.J., Heath, J.K., Cooper, H.J.: FAIMS and phosphoproteomics of fibroblast growth factor signaling: enhanced identification of multiply phosphorylated peptides. J. Proteome Res. 14, 5077–5087 (2015)
Pfammatter, S., Bonneil, E., Thibault, P.: Improvement of quantitative measurements in multiplex proteomics using high-field asymmetric waveform spectrometry. J. Proteome Res. 15, 4653–4665 (2016)
McManus, F.P., Lamoliatte, F., Thibault, P.: Identification of cross talk between SUMOylation and ubiquitylation using a sequential peptide immunopurification approach. Nat. Protoc. 12, 2342–2358 (2017)
Prasad, S., Belford, M.W., Dunyach, J.J., Purves, R.W.: On an aerodynamic mechanism to enhance ion transmission and sensitivity of FAIMS for nano-electrospray mass spectrometry. J. Am. Soc. Mass Spectrom. 25, 2143–2153 (2014)
Barnett, D.A., Ouellette, R.J.: Elimination of the helium requirement in high-field asymmetric waveform ion mobility spectrometry (FAIMS): beneficial effects of decreasing the analyzer gap width on peptide analysis. Rapid Commun. Mass Spectrom. 25, 1959–1971 (2011)
Enserink, J.M.: Sumo and the cellular stress response. Cell Div. 10, 4 (2015)
Golebiowski, F., Matic, I., Tatham, M.H., Cole, C., Yin, Y., Nakamura, A., Cox, J., Barton, G.J., Mann, M., Hay, R.T.: System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2, ra24 (2009)
Bonneil, E., Pfammatter, S., Thibault, P.: Enhancement of mass spectrometry performances for proteomics analyses using high-field asymmetric waveform spectrometry (FAIMS). J. Mass Spectrom. 50, 1181–1195 (2015)
Tyanova, S., Temu, T., Cox, J.: The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protocols. 11, 2301–2319 (2016)
Gill, G.: Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr. Opin. Genet. Dev. 13, 108–113 (2003)
Seo, J., Lee, K.J.: Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. J. Biochem. Mol. Biol. 37, 35–44 (2004)
Filtz, T.M., Vogel, W.K., Leid, M.: Regulation of transcription factor activity by interconnected post-translational modifications. Trends Pharmacol. Sci. 35, 76–85 (2014)
Rosonina, E., Akhter, A., Dou, Y., Babu, J., Sri Theivakadadcham, V.S.: Regulation of transcription factors by sumoylation. Transcription. e1311829 (2017)
Hong, Y., Rogers, R., Matunis, M.J., Mayhew, C.N., Goodson, M.L., Park-Sarge, O.K., Sarge, K.D.: Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 276, 40263–40267 (2001)
Fulda, S., Gorman, A.M., Hori, O., Samali, A.: Cellular stress responses: cell survival and cell death. International Journal of Cell Biology. 2010(23), (2010)
Roy, A.L.: Biochemistry and biology of the inducible multifunctional transcription factor TFII-I: 10 years later. Gene. 492, 32–41 (2012)
Chu, Y., Yang, X.: SUMO E3 ligase activity of TRIM proteins. Oncogene. 30, 1108–1116 (2011)
Guo, L., Giasson, B.I., Glavis-Bloom, A., Brewer, M.D., Shorter, J., Gitler, A.D., Yang, X.: A cellular system that degrades misfolded proteins and protects against neurodegeneration. Mol. Cell. 55, 15–30 (2014)
Martin, N., Schwamborn, K., Schreiber, V., Werner, A., Guillier, C., Zhang, X.D., Bischof, O., Seeler, J.S., Dejean, A.: PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. EMBO J. 28, 3534–3548 (2009)
Luo, X., Kraus, W.L.: On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 26, 417–432 (2012)
Woodhouse, B.C., Dianov, G.L.: Poly ADP-ribose polymerase-1: an international molecule of mystery. DNA repair. 7, 1077–1086 (2008)
Morris, J.R., Boutell, C., Keppler, M., Densham, R., Weekes, D., Alamshah, A., Butler, L., Galanty, Y., Pangon, L., Kiuchi, T., Ng, T., Solomon, E.: The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 462, 886–890 (2009)
Takahashi, A., Matsumoto, H., Nagayama, K., Kitano, M., Hirose, S., Tanaka, H., Mori, E., Yamakawa, N., Yasumoto, J., Yuki, K., Ohnishi, K., Ohnishi, T.: Evidence for the involvement of double-strand breaks in heat-induced cell killing. Cancer Res. 64, 8839–8845 (2004)
Takahashi, A., Mori, E., Somakos, G.I., Ohnishi, K., Ohnishi, T.: Heat induces gammaH2AX foci formation in mammalian cells. Mutat. Res. 656, 88–92 (2008)
Cheng, J., Wang, D., Wang, Z., Yeh, E.T.: SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol. Cell. Biol. 24, 6021–6028 (2004)
David, G., Neptune, M.A., DePinho, R.A.: SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J. Biol. Chem. 277, 23658–23663 (2002)
Acknowledgements
The authors thank Jean-Jacques Dunyach, Michael Belford and Satendra Prasad (Thermo Fisher Scientific) for valuable help and assistance with the FAIMS interface. The Institute for Research in Immunology and Cancer (IRIC) receives infrastructure support from IRICoR, the Canadian Foundation for Innovation, and the Fonds de Recherche du Québec-Santé (FRQS). IRIC proteomics facility is a Genomics Technology platform funded in part by the Canadian Government through Genome Canada.
Funding
This work was carried out with financial support from the Natural Sciences and Engineering Research Council (NSERC 311598) and the Genomic Applications Partnership Program (GAPP) of Genome Canada. The Institute for Research in Immunology and Cancer (IRIC) receives infrastructure support from IRICoR, the Canadian Foundation for Innovation, and the Fonds de Recherche du Québec - Santé (FRQS). IRIC proteomics facility is a Genomics Technology platform funded in part by the Canadian Government through Genome Canada.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pfammatter, S., Bonneil, E., McManus, F.P. et al. Gas-Phase Enrichment of Multiply Charged Peptide Ions by Differential Ion Mobility Extend the Comprehensiveness of SUMO Proteome Analyses. J. Am. Soc. Mass Spectrom. 29, 1111–1124 (2018). https://doi.org/10.1007/s13361-018-1917-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-018-1917-y