Abstract
Roundabout 1 (Robo1) interacts with its receptor Slit to regulate axon guidance, axon branching, and dendritic development in the nervous system and to regulate morphogenesis and many cell functions in the nonneuronal tissues. This interaction is known to be critically regulated by heparan sulfate (HS). Previous studies suggest that HS is required to promote the binding of Robo1 to Slit to form the minimal signaling complex, but the molecular details and the structural requirements of HS for this interaction are still unclear. Here, we describe the application of traveling wave ion mobility spectrometry (TWIMS) to study the conformational details of the Robo1-HS interaction. The results suggest that Robo1 exists in two conformations that differ by their compactness and capability to interact with HS. The results also suggest that the highly flexible interdomain hinge region connecting the Ig1 and Ig2 domains of Robo1 plays an important functional role in promoting the Robo1-Slit interaction. Moreover, variations in the sulfation pattern and size of HS were found to affect its binding affinity and selectivity to interact with different conformations of Robo1. Both MS measurements and CIU experiments show that the Robo1-HS interaction requires the presence of a specific size and pattern of modification of HS. Furthermore, the effect of N-glycosylation on the conformation of Robo1 and its binding modes with HS is reported.

ᅟ
Similar content being viewed by others
Introduction
Robo-Slit signaling is one of the best studied axon guidance pathways and their interaction is known to be critically modulated by heparan sulfate (HS). For instance, during the assembly of the central nervous system (CNS), commissural axon migration at the CNS midline is regulated by the interactions between axon guidance molecules and their receptors, such as the interaction between attractive cues Netrin and its receptor Deleted in colorectal carcinoma, which stimulates pre-crossing of axons to the contralateral side [1]. Interaction between repelling cues Slit and its cellular roundabout receptor (Robo) impedes aberrant post-crossing of ipsilateral axons [2, 3]. Though best known for its roles in neuron development and axon pathfinding, Robo-Slit signaling is not confined to the nervous tissue and has been implicated in multiple nonneuronal tissues, contributing to a variety of development and physiological processes through mediating cell proliferation [4], cell adhesion [5], cell migration and mobility [6, 7], angiogenesis [8], as well as dendritic development and branching [9]. Robo-Slit signaling has also been associated with progression [10, 11], metastasis [12], and angiogenesis of tumor [13]. Deletion, epigenetic modification, and abnormal expression of Robo and Slit genes occurred in different types and different stages of cancers [14, 15], making them attractive candidates for cancer diagnostics and therapeutics.
The Robos are a family of transmembrane proteins that are mainly expressed in CNS neurons. Sharing structural motifs with the Ig superfamily of cell adhesion molecules (CAM) [16], Robos are composed of five Ig-like domains and three fibronectin type III motifs (FN3) in its extracellular region (except for Robo4), followed by a transmembrane region and an intracellular cytoplasmic domain. Vertebrates have four types of Robo (Robo1–4). Though they have distinct cytoplasmic domains, their Ig1 and Ig2 domains are highly conserved and are believed to be responsible for mediating the Robo-Slit interaction [17].
Two crystal structures are available for the two N-terminal Ig domains of human Robo1 [18]. In the first crystal structure, Ig1 and Ig2 are aligned in a linear format, with the final strand of Ig1 extending and forming a 100-Å-long interdomain region to connect the two domains. Compared with the first crystal form, the Ig2 of the second crystal form is rotated by 39.9° at the hinge region, resulting in a slightly bent arrangement. Two similar crystal structures have been observed for the first two Ig domains of Drosophila Robo1 [19]. One tetragonal crystal form and one monoclinic crystal form were observed, corresponding to the slightly bent and linear crystal structures for human Robo1, respectively. Both of the previous studies on human and Drosophila Robo1 observed substantial flexibility embedded in the interdomain hinge region and raised questions about whether this flexibility is related to the biological function of Robo1.
Both Robo1 and Slit are HS binding proteins. HSs are linear, highly sulfated glycosaminoglycan (GAG) polysaccharides. The backbone structure of HS is a product of polymerization of disaccharide units containing glucosamine (GlcN) and uronic acid residues (glucuronic acid (GlcA) or iduronic acid (IdoA)) linked through a 1–4 glycosidic linkage. Modification of HS backbone including C5 epimerization of uronic acid, deacetylation of glucosamine, and addition of various types of N- or O-sulfation generate enormous sequence heterogeneity. HSs interact with many proteins and play important roles in a variety of physiological and pathological processes by regulating the structure, conformation, distribution, stability, and activity of these proteins [20, 21].
Substantial in vivo and in vitro studies suggest that HS plays an indispensable role in Robo-Slit-mediated axon guidance: null mutations in Caenorhabditis elegans enzymes that are responsible for HS modifications including sulfotransferases and epimerases caused defected axonal and cellular guidance [22]; mutations in Drosophila syndecan altered the Slit distribution in developing axons [23] and interfered with the slit signal transmission in target cells [24]; enzymatic digestion of endogenous HS using heparinase III destabilized Robo1-Slit interaction and abolished the repulsive activity of Slit [25]. Collectively, HS has dual roles in Robo1-Slit signaling: the main role of HS is to act as co-receptors of Slit, simultaneously interacting with the LRR2 domain of Slit and Ig1–2 domains of Robo1, enhancing and stabilizing the Robo1-Slit interaction by increasing their binding affinity by tenfold and promoting the formation of trimeric Robo1-Slit-HS signaling complexes [26, 27]. An additional function of HS is to mediate the distribution of Slit through mechanism of either active transport, buffered diffusion, or selective stabilization [23], therefore defining the concentration and accessibility of Slit on the extracellular surface [28]. However, more experimental evidence is required to determine whether the HS-dependence of Robo-Slit interaction is required in all cell types and tissue types in mammals [29].
Though the significance of HS in Robo-Slit interaction has been well established, a clear elucidation of all the molecular details of this interaction is still missing. Previous mutagenesis and X-ray crystallography experiments identified the binding site of HS as a contiguous basic patch spanning both the surfaces of Robo1 and Slit, but the structural requirements of the bound HS have not been well defined. Also, more experiments are required to elucidate the molecular mechanism of how exactly the interaction between Robo and Slit is programmed into signal transduction through interacting with HS.
Previously, we developed and applied traveling wave ion mobility spectrometry (TWIMS) to examine the interaction between antithrombin III (ATIII) and heparin [30] as well as the interaction between fibroblast growth factor 1 (FGF1) and HS [31]. TWIMS is capable of providing conformational and stoichiometric details that are consistent with the known behaviors of these two well-characterized interactions. We also observed that the IM profiles of GAG-protein complexes provide useful data regarding the binding specificity of their interactions. In this current study, TWIMS is used to investigate the less well-characterized Robo1-HS interaction. Only the first two Ig domains were used as a representative of intact Robo1 since they are the most conserved regions of Robo1 and participate in the binding of Robo1 and Slit with HS [32, 33]. The results provide new insights into the functional significance of the conformational flexibility of the Ig1–2 domains of Robo1, and how HS implements its biological function through selectively recognizing and interacting with different conformations of the Ig1–2 domains of Robo1. The results also provide an in-depth understanding of the way HS modulates its functional interaction with Robo1 through its fine structure. Moreover, a new mechanism for the activation of Robo1 by HS and the promotion of the Robo1-HS interaction is proposed.
Experimental
Reagents
All chemicals and solvents (ammonium acetate, methanol, water, and formic acid) were of HPLC grade and purchased from Sigma-Aldrich, (St. Louis, MO). Protein calibrants (myoglobin from equine heart, cytochrome c from equine heart, avidin from egg white, concanavalin A from Canavalia ensiformis, and bovine serum albumin) were purchased from Sigma-Aldrich. HS hexasaccharides (dp6) to dodecasaccharides (dp12) were chemo-enzymatically synthesized as previously described [34]. HS tetrasaccharide (dp4) was chemically synthesized as previously described by fluorous supported modular synthesis [35].
Protein Expression
An expression vector encoding the human roundabout homolog 1 precursor (Robo1, NP_002932, Uniprot Q9Y6N7) Ig domains 1 and 2 (residues 58–266) in the pGEn2 vector was previously described [36]. The vector encodes a fusion protein with an NH2-terminal signal sequence followed by an 8xHis tag, AviTag [37], “super-folder” GFP [38], the TEV protease recognition site [39], and the Ig1–2 domains of Robo1. Expression of the construct in mammalian cells leads to translocation of the fusion protein into the secretory pathway, glycosylation, and secretion as a GFP-Robo1 Ig1–2 fusion into the conditioned media.
A separate construct encoding leukocyte common antigen-related receptor protein tyrosine phosphatases (LAR-RPTP, NM_002840, Uniprot P10586) NH2-terminal signal sequence and Ig domains 1 and 2 (residues 1–231) in the pGEc2 vector. This construct encodes a fusion protein containing the LAR signal sequence and Ig1–2 domains followed by the TEV protease recognition site, “super-folder” GFP, AviTag, and 8xHis tag. Expression of the construct in mammalian cells leads to translocation of fusion protein into the secretory pathway and secretion as a Robo1 Ig1–2-GFP fusion into the conditioned media. Mass spectrometry data indicated that the single Asn117-Thr-Ser119 N-glycosylation sequon in the LAR coding region is not occupied with an N-glycan structure.
Both fusion protein expression constructs were transiently transfected into mammalian cells using either FreeStyleTM 293-F cells (Thermo Fisher Scientific) for protein production containing wild-type glycans (complex-type glycans) or HEK293S (GnTI-) cells [40] (ATCC) for expression with high mannose glycan structures (predominately Man5GlcNAc2-Asn glycans). Both cell lines were maintained in culture media comprised of 9 volumes FreeStyleTM 293 expression media (Thermo Fisher Scientific) and 1 volume EX-CELL® 293 serum-free medium (Sigma) (9:1 media) as previously described. Suspension cultures (1 L) of either HEK293S (GnTI-) cells or FreeStyleTM 293-F cells were transfected with the respective plasmid DNAs using polyethyleneimine (linear 25 kDa PEI, Polysciences) as transfection reagent as previously described [41]. The cultures were diluted 1:1 with culture medium containing 4.4 mM valproic acid (2.2 mM final concentration) 24 h after transfection, and protein production was continued for a further 5 days at 37 °C.
Protein Purification
Protein purification, deglycosylation, and tag removal employed workflows similar to prior structural studies on rat ST6GAL1 [41]. Briefly, the conditioned culture medium was loaded on a Ni2+-NTA Superflow (QIAGEN) column equilibrated with 20 mM HEPES, 300 mM NaCl, 20 mM imidazole, pH 7.4, washed with column buffer, and eluted successively with column buffers containing stepwise increasing imidazole concentrations (40–300 mM). The eluted fusion protein was pooled, concentrated, and mixed with recombinant tobacco etch virus (TEV) protease (1:10 ratio relative to GFP-Robo1 or LAR-GFP) and incubated at 4 °C for 24 h. The samples were diluted 15-fold in elution buffer without imidazole and re-applied to the Ni2+-NTA column to allow collection of the Robo1 or LAR products in the flow-through fractions. To produce Robo1 with minimal glycosylation, the purified protein expressed in HEK293S(GnTI-) cells was treated with endoglycosidase F1 (EndoF1), which cleaves the glycan structure between the two core GlcNAc residues and truncates glycans to a single GlcNAc residue [41]. EndoF1 was added concurrently with TEV protease at a 1:10 ratio relative to the GFP-Robo1 and incubated at 4 °C for 24 h. Dilution to lower the imidazole concentration was followed by passing through a Ni2+-NTA column to remove the fusion tag and His-tagged TEV protease and EndoF1 to allow collection of the Robo1 product in the flow-through fractions. Each of the protein forms (Robo1 containing wild-type glycans, Man5GlcNAc2-Asn glycans, or a single GlcNAc, or LAR containing no glycans) were further purified on a Superdex 75 gel filtration column (GE Healthcare) and peak fractions were collected and concentrated by ultrafiltration to 30 mg/mL for mass spectrometry analysis.
Sample Preparation
For mass spectrometry (MS) analyses under nondenaturing conditions, Robo1 was diluted in 20 mM ammonium acetate buffer, pH 6.8, to a final concentration of 20 μM. Robo1-HS complex was obtained by incubating Robo1 with HS oligosaccharides at room temperature for 60 min. Protein calibrants were diluted in either denaturing solution or nondenaturing solution to a final concentration of 10 μM.
IMMS Measurement and Data Analysis
NanoESI-IMMS experiments were performed using a quadrupole-TWIMS-TOF hybrid mass spectrometer (Synapt G2 HDMS; Waters Corp., Manchester, UK) in positive ionization mode. Protein samples were injected into the nanoESI source through a fused-silica emitter (PicoTip; New Objective, Woburn, MA, USA) with a flow rate varying from 0.2 to 0.5 μL/min. The applied experimental parameters were capillary voltage, 1.4 kV; sampling cone voltage, 20 V; extraction cone voltage, 5 V; source temperature, 30 °C; flow rate of nitrogen in the IM ion guide, 50 mL/min; flow rate of helium in the helium cell, 180 mL/min; and transfer collision energy, 0 V. Different sets of wave height and corresponding wave velocity were examined to optimize the mobility separation. The drift times of the calibrants and Robo1 samples were measured and identical experimental conditions were stringently applied.
For monitoring the collisional-induced unfolding (CIU) of Robo1 and its complexes, protein ions of a selected charge state were isolated in a quadrupole mass filter and activated in the trap ion guide where subsequent collisional-induced activation of ions takes place, followed by separation in the TWIMS ion mobility stage, and detection by TOF-MS.
Data analysis was performed using MassLynx 4.1 and Driftscope (Waters Corp., Manchester, UK). The CIU data was analyzed using CIUSuite [42]. CIU fingerprints of protein ions, which records the relative ion intensity (normalized and smoothed using Savitsky-Golay filter) as a function of collision energy and drift times, were shown in a 2D contour plot using the CIUSuite plot function. The CIUsuite detect function was applied to identify and extract the most intense CIU features of the fingerprint based on a first derivative analysis, providing centroid drift times, transitional voltage, and stability range of each detected feature.
Converting Drift Times into Collision Cross Sections
The collision cross sections (CCSs) of Robo1 and Robo1-HS complexes were calibrated based on an empirical relationship between the drift times of protein calibrants and their known CCSs obtained previously by DTIMS. Briefly, a selected set of native and denatured protein calibrants, with a mass range from 12 to 102 kDa and a CCS range from 2303 to 5550 Å2 were employed. The drift times of these calibrates were corrected for mass-dependent flight time spent in the transfer ion guide and TOF mass analyzer and mass-independent flight time spent in the transfer ion guide. The CCSs of these calibrants were corrected for their charge state and reduced mass with respect to the buffer gas. The natural logarithm of corrected CCSs were plotted against the natural logarithm of corrected drift times and a mathematical formula (Ln Ω′ = A × Ln dt′ + B ) was derived. The coefficient A was extracted to calculate the effective drift times dt′′: dt′′ = \( {\left({\mathrm{dt}}^{\prime}\right)}^A\frac{z}{\sqrt{\mu }} \). A calibration curve was generated by plotting the literature CCSs as a function of dt′′, as shown in the Supplementary figure 1. The experimental CCS of the analyte ion was derived from this calibration curve based on the measured drift time [43].
Molecular Dynamics Simulations and Theoretical CCS Calculations
A model of the Robo-HS complex was created by extending the glycan within a model previously produced according to NMR-based restraints [44], which was in turn built using a crystal structure of the human Robo Ig1 and Ig2 (PDB ID: 2V9R) [18]. The ligand was extended with Chimera [45] by aligning GLYCAM-minimized disaccharide units to the end of the tetrasaccharide, producing a final hexasaccharide sequence of (IdoA2S-α-(1–4)-GlcNS6S)3. An additional “bent” model of Robo was created by separating the Ig1 and Ig2 domains within the Robo structure at residue A109 (PDB ID: 2V9R) and aligning each to the Ig domains of an LAR structure (PDB ID: 2YD5) with Chimera. The linker regions between Ig domains (residues E107-Q117) was then refined with the Chimera interface with MODELLER [46]. Glycosylated models of both Robo forms were created via the GLYCAM Web tool (www.glycam.org).
Topology and coordinate files for Robo1 and Robo1-HS complexes were generated using the tLeap program, employing the ff99SB [47] and GLYCAM06 (version j) [48] parameters for the protein and GAGs, respectively. Each system underwent energy minimization (1000 steps) in implicit solvent (IGB = 2). The net charge on each system, after energy minimization, was neutralized by addition of an appropriate number of counter ions (Na+ or Cl−). This was followed by solvation with TIP3P water molecules in a cubic box extending at least 12 Å from any atom of the solute.
All molecular dynamic (MD) simulations were performed with the GPU implementation of pmemd, pmemd.cuda in Amber14 [49, 50]. Energy minimization of the solvent was performed in an NVT ensemble (500 steps of steepest descent and 20,000 steps of conjugate gradient), followed by a full system energy minimization (1000 steps of steepest descent and 25,000 steps of conjugate gradient). The systems were heated from 5 to 300 K over 60 ps in an NVT ensemble, with a weak positional restraint (10 kcal/mol-Å2) on the atoms in the solute. A Berendsen-type thermostat with a time coupling constant of 1 ps was utilized for temperature regulation. Equilibration and production were performed at constant pressure (NPT ensemble; 1 atm) with a pressure relaxation time of 1 ps. Covalent bonds involving hydrogen atoms were constrained using the SHAKE algorithm, allowing a simulation time step of 2 fs. Scaling factors for 1–4 interactions were set to the recommended values of 1.0 and 1.2 for the GAG and protein, respectively, and a non-bonded interaction cutoff of 8.0 Å was employed. Long-range electrostatics were computed with the particle mesh Ewald (PME) method [51]. The system equilibrated at 300 K for 1 ns before a production run that lasted 100 ns.
Post-processing of the MD simulations was performed with the CPPTRAJ module of AmberTools14 [52]. CCSs were calculated using 50 frames extracted from the simulation using MOBCAL. Each of these frames was minimized in vacuo, followed by the calculation of CCS using the projection approximation and trajectory methods. Graphical representations were generated using VMD [53]. Cartoon representations of glycans follow the SNFG format [54], and the 3D shapes were created with the 3D–SNFG plugin for VMD [55].
Results and Discussion
Native Mass Spectrometry of Unbound and HS-Bound Ig1–2 Domains of Robo1
The native mass spectrum of the N-terminal Ig1–2 domains of unbound Robo1 was first acquired under non-denaturing conditions without the addition of any HS (Figure 1a). A dominant monomeric form of Robo1 with a charge state distribution ranging from +13 to +9 over a m/z range of 1800–2700 was observed. The narrow distribution of these lower charge states indicates that Robo1 ions adapt a natively folded and compact conformation with fewer basic sites exposed for protonation, and the solution structure of Robo1 survived the ionization process and the environment of the TWIMS instrument. An average molecular weight of approximately 23.5 kDa was obtained, in agreement with the literature molecular weight of Robo1 with a single N-acetylglucosamine (GlcNAc) residues attached at asparagine (210). A dimeric form of Robo1 with a charge state distribution ranging from +16 to +13 was also observed, but its biological relevance is unclear due to its low abundance.
(a) Native ESI mass spectra of unbound Robo1, which shows a dominant monomeric form of Robo1 with a narrow charge state distribution. (b) Robo1 incubated with an equi-molar mixture of HSs ranging in size from dp4 to dp12. Complexes for the +12, +11, and +10 charge states are indicated by color code that indicates the length (dp) of the bound HS. For all three charge states, dp8 (indicated by a green square) is the preferred size for binding
Robo1 was next incubated with an equi-molar mixture of fully sulfated HS ranging in size from dp4 to dp12 to reach a final molar ratio of 1:3 (protein to GAG). These HS oligosaccharides consist of repeating disaccharide units of (IdoA2S-α-(1–4)-GlcNS6S)n. The overall charge state distribution remained unchanged, while additional peaks corresponding to the formation of Robo1-HS complexes in a binding stoichiometry of 1:1 were observed for each charge state (Figure 1b). The fact that HS as short as a dp4 can be able to form 1:1 complex with Robo1 is in accordance with a previous SPR competition experiment which showed that HSs longer than dp2 is required to efficiently compete with chip immobilized heparin for interacting with Robo1 [56]. The absence of complexes in other binding stoichiometries agreed with previous findings determined by size-exclusion chromatography [26].
Size-Dependent Specificity of Robo1-HS Interactions
Since HS oligosaccharides of different sizes competed with each other for Robo1, the abundance of the complex peaks should provide an experimental estimation of their binding affinity toward Robo1. Despite a few overlapping peaks between +n charge state of unbound Robo1 and +(n + 1) charge state of Robo1-HS complex, the data in Figure 1 show that for all the charge states, the Robo1-dp8 complex is the most abundant peak among all the complexes, and its abundance is even higher than those complexes with longer HSs that carry more charged modifications. These observations clearly indicated that Robo1-HS interaction is highly sensitive to the size of HS instead of driven by overall level of charge/sulfation. The facts that a high affinity interaction between Robo1 and HS requires a structural motif of a distinctive size, and adding more saccharide units or sulfations inhibits the interaction agree with the binding behavior of a highly specific interaction.
Coincidently, a previous experiment on exposing Xenopus embryonic brains to selectively desulfated heparins followed by assessing their mistargeting-inducing activity suggest that the minimum size of HS required for mistargeting is dp8 [57]. Another previous study based on separation and sequencing of Robo1-bound, natural derived HSs also identified a dp8 as a potential ligand for this interaction, despite that the detailed sulfation patterns are different from the dp8 used in this study [58]. Taken together with our observation that a HS dp8 has the highest binding affinity toward Robo1 when competing with longer, more sulfated HSs suggest that the binding epitope on Robo1 could be a shallow grove which prefers to accommodate a HS dp8.
Conformational Change Involved in Robo1-HS Interaction
Evidence of a possible conformational change within Robo1 was observed upon the binding of HS/Hp in previous studies: using surface plasmon resonance (SPR), a much better fit for the Robo1-Heparin/HS binding data was achieved with a two-state (conformational change) binding model than a 1:1 L model [56]; using hydroxyl radical protein footprinting (HRPF) mass spectrometry (MS), residues on Ig2 domains were observed to be less solvent-exposed upon the binding of heparin, suggesting a dynamic movement of Ig2 domain or a conformational change of the first two Ig domains [59]. Therefore, in the current study, TWIMS was applied to investigate whether Robo1 undergoes a conformational change due to the binding of HS and to directly measure the conformational change if there is any.
The arrival time distribution of unbound Robo1 (+11) displayed a distribution featuring two separate peaks. Two Gaussian distributions were used to fit the measured arrival time distribution according to its shape, with each Gaussian distribution representing a potentially stabilized gas-phase conformation (Figure 2a). The conformation with a lower drift time corresponds to a more compact conformation of Robo1, with a relative peak abundance of 20.1 ± 0.2%. The more dominant conformation with a higher drift times corresponds to a more open or extended conformation. The arrival time distributions of lower charge states, +10 and +9 favored the more compact conformation of the protein (Supplementary figure 2). Lower charge states often correlate to more native-like structures for gas-phase proteins, and these data suggest that the more compact form of the protein may be more native-like and therefore of greater biological relevance.
Arrival time distributions (ms) of +11 charge state of unbound Robo1 (a) and Robo1 bound with a fully sulfated HS dp6 (b) both display one compact conformation and one extended conformation. The relative ion population areas of the more compact conformation for unbound and complexed Robo1 are 20.1 ± 0.2 and 51.7 ± 1.1%, respectively, indicating that HS dp6 preferentially interacts with the more compact conformation
The arrival time distribution of Robo1 (+11) in complex with a fully sulfated HS dp6 (Figure 2b) also exhibited two peaks, corresponding to two stabilized gas-phase conformations with different shape and compactness. The relative peak area of the conformation with a lower drift time is 51.7 ± 1.1%, indicating a higher population of the compact conformation for Robo1_HS complex. The lower charge states (+10 and +9) only show a single conformation, corresponding to the more compact structure, as well (Supplementary figure 2). Different incubation time ranging from 0.5 to 2 h was tested to evaluate the kinetic stabilities of the observed structural distributions. As shown in Supplementary figure 3, the more compact conformation remained the most abundant conformation regardless of the incubation time. We attributed the dramatically shifted conformation equilibrium upon the binding of HS to a more favorable interaction between HS and Robo1 in the more compact conformation.
The experimental collision cross sections (CCSs) for the two conformations observed for both unbound and HS-bound Robo1 were calibrated based on the measured drift times and are summarized in Table 1. The experimental CCS of the compact conformation is approximately 17.3% smaller than the other conformation in the unbound form, which is in close agreement with the experimental CCS difference (approximately 16.8%) between the two conformations in the complexed form of Robo1.
Previously, two distinctive crystal forms including one linear shape and one slightly bent shape were resolved for the Ig1–2 region of human Robo1 [18]. Both two crystal forms were used for the calculation of theoretical CCSs of Robo1, which were then compared with the experimental CCSs. Though the crystal structure of HS-bound Robo1 is not available, a constrained docking model proposed by a previous NMR study was used to estimate the theoretical CCS of the Robo1-HS complex [44]. Three methods were used for calculating theoretical CCSs, as shown in Table 2. Previous studies showed that projection approximation (PA) and trajectory (TM) method set the lower and upper limit of the range of acceptable experimental CCSs, respectively, while calibrating the PA CCSs to generate scaled PA CCSs allows a better comparison of experimental and theoretical CCSs [60, 61].
To our surprise, though the elbow angle of the two crystal forms varies from 140° to 180°, their theoretical CCSs are quite comparable. It is also worth noting that the theoretical CCSs evaluated from the crystals structures are in close agreement with the experimental CCS of the more extended conformation. These observations indicated that in addition to the conformation that matches with the crystal structures, a new conformation was observed in TWIMS experiment, corresponding to a more compact structure of Robo1 with a much smaller CCS. The fact that HS dp6 preferentially interacts with this new and more compact conformation suggests a higher biological relevance of the new conformation, though solution-phase evidence is necessary to confirm this suggested postulation.
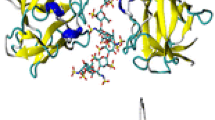
A putative model for the newly identified conformation was proposed, based on the structural similarities of the extracellular Ig1–2 domains of leukocyte common antigen-related receptor protein tyrosine phosphatase (LAR-RPTP). LAR-RPTP also belongs to the cell adhesion family of proteins with GAG binding affinity and is also functionally involved in the development of vertebrate nervous system [62]. In its crystal structure, the first two Ig domains of LAR-RPTP align parallel with each other and form a much more dramatically bent, V-shaped structure [63], which was used as the template to build a LAR-Robo1 hybrid model (Figure 3). The theoretical CCS of the proposed LAR-Robo1 hybrid model was measured, and is in close agreement with the experimental CCS of the more compact conformation of Robo1 (Table 2), indicating that this LAR-Robo1 hybrid model may represent a similar compact structure that Robo1 adopted in TWIMS experiment.
The proposed LAR-Robo1 hybrid model for predicting the structure of the compact conformation of Robo1. The Ig1 domain of Robo1 is shown in orange and the Ig2 domain is shown in purple, with the attached glycan shown in 3D–SNFG representation. The structure of LAR-RPTP which was used as the template is shown in gray
We have also analyzed the Ig1–2 domains of LAR-RPTP. In contrast to Robo1, only one compact conformation was observed for LAR-RPTP. The observance of a single conformation is expected, since the V-shaped structure of LAR-RPTP is greatly stabilized through the presence of tight interaction including hydrogen bonds, salt bridges, and hydrophobic interactions between its Ig1 and Ig2 domains. Theoretical CCSs of LAR-RPTP also agreed with its experimental CCSs (data not shown).
Although the role of HS in the Robo1-Slit signaling is well recognized, the molecular details of how they interact to form the ternary signaling complex are still unknown. Previous experiment that selectively mutated amino acids from different domains of Robo1 followed by evaluating the effect using solid-phase assay identified that the major Slit binding site is primarily conserved in Ig1 domain of Robo1 [19]. Using SPR, a ten times lower Kd was measured for the Ig2 domain, indicating a negligible binding affinity compared with Ig1 [18]. However, a previous domain deletion mutagenesis experiment showed that removing the Ig2 domain completely abolished the biological activity of Robo1 to interact with Slit in vitro [32]. A recent domain deletion study suggests that though Ig1 domain played a critical role in Slit binding, Ig2 domain is the only Ig domain among Ig2–5 domains that contributed to Robo1-slit binding as well as axonal localization guidance and repulsive signaling in vivo [64]. Therefore, it seems that both the Ig1–2 domains of Robo1 are required for interacting with Slit, but this also contradicts with the crystallographic findings of Robo1-Slit complex, which suggest that a conserved motif on Slit forms a concave binding site for Robo1 that can only accommodate a single Ig domain [18]. These observations raised questions about the contribution of Ig2 domain in the Robo1-Slit interaction. Moreover, studies on the crystal structures of Ig1–2 domains of human and Drosophila Robo1 both reported substantial flexibility of the hinge region, and have both raised questions about the biological relevance of this flexibility.
Taking our and other’s observations together, we proposed that HS plays an essential role in activating Robo1. Native Robo1 prefers to stay in a more extended conformation, as shown in our TWIMS measurement and identified by previous X-ray crystallography experiments. The function of HS is to either selectively recognize and interact with the less abundant compact conformation of Robo1 or induce a conformational change of the more extended conformation as they interact with each other, resulting in the increased population of the more compact conformation in the complexed form of Robo1. It is likely that the compact conformation of Robo1 has higher biological activity than the extended conformation and is more efficient in stabilizing the interactions between Slit and Robo1, further facilitating the formation of the ternary Robo1-Slit-HS complex and the downstream signaling processes.
Sulfation Pattern-Dependent Specificity of Robo1-HS Interaction
In an effort to examine the effect of structural diversity on Robo1-HS interactions, an equi-molar mixture of Hp dp12 with different sulfation patterns were incubated with Robo1 to reach a final molar ratio of 1:2 (protein to GAG). The structures of the dp12 oligomers are shown in the supplementary data. Peaks corresponding to the formation of 1:1 Robo1-HS dp12 complexes were observed for the fully sulfated, N, 6-O-sulfated, and N, 2-O-sulfated, whereas no complex corresponding to the N-sulfated dp12 bound Robo1 was observed (Figure 4). It is evident that the peak abundance for fully sulfated dp12-bound Robo1 is much lower than those formed with HS missing 6-O or 2-O sulfation. The fact that removing of 2-O or 6-O sulfation improved binding stands in great contrast to the less specific FGF1-HS binding system that we investigated before [31]. These observations indicated that Robo1-HS interaction is sensitive to the sulfation pattern of HS rather than driven by the amount and density of charged groups. However, since the peak abundance of the N, 6-O-sulfated dp12-bound complex and the N, 2-O-sulfated dp12-bound complex are comparable, we cannot compare the contribution of 2-O sulfation and 6-O sulfation based on MS measurements alone.
We then checked the ion mobility behavior of Robo1-HS complex with different modified HS dp12s (Figure 5). The IM profiles of different Robo1 complexes were quite distinct from each other, indicating that the sulfation pattern of HS dictates different binding affinity toward the two conformations of Robo1. Specifically, the compact form of Robo1 has a discernable preference for N-S, 6-S modification on the amino sugar. 2-S modification of the uronic acid seems to reduce this preference.
Arrival time distributions (ms) of +11 charge state of Robo1 in complex with fully sulfated dp12 (upper), Robo1 with N, 2-O-sulfated dp12 (middle), and Robo1 with N, 6-O-sulfated dp12 (bottom). The relative ion population areas of the compact conformation for each Robo1-dp12 complex are 43.5 ± 0.7, 45.7 ± 0.7, and 54.1 ± 0.4%, respectively. The observation that HSs with different sulfation patterns have different preference for these two conformations indicates HS sequence specificity
A previous SPR study that measured the competition between chip immobilize heparin and selectively desulfated heparin-derived oligosaccharides in solution demonstrates that N-sulfation and 6-O sulfation contribute more to Robo1-Slit interaction than 2-O sulfation [56]. Another study which measured the binding affinity of chip immobilized Robo1 and a series of chemically modified HS dp4 suggests that 2-O sulfation have a negative effect on the interaction [58]. Comparing the IM profiles of fully sulfated dp12-bound complex and de-2-O-sulfated dp12-bound complex, the relative ion population area of the more compact conformation of Robo1 increased by over 10% after removing 2-O sulfation. This observation indicates that removing 2-O sulfation led to a more effective binding with the compact conformation, consistent with those previous results. Taken together, Robo1-HS interaction requires a specific combination of selective patterns of sulfation for recognizing the more functionally relevant conformation of Robo1.
This observation is in great contrast to the behavior of the chemokine MCP-1 dimer-HS interaction [65]. Though two conformations were also observed for MCP-1 dimer, the relative ion population areas of the two conformations of MCP-1 dimer in complex with differently modified HSs were quite comparable. Only fully sulfated HS demonstrated a preferential binding toward the compact conformation, whereas selectively desulfated HS showed decreased binding affinity toward the more compact conformation. The different behavior of Robo1 and MCP-1 indicates that Robo1 has a more stringent and more specific requirement for its HS ligand.
The Effect of N-Glycosylation on Conformation and Binding Affinity of Robo1
Native Robo1 has a glycosylation site at the asparagine residue located near the C-terminus of its Ig1 domain, with a full size, complex type of N-glycan attached. The results above were detained with a more homogeneous form of Robo1 with a single GlcNAc residue attached. Additional experiments were performed to investigate whether different size and composition of N-glycan may affect the form, population/equilibration, and stability of the gas-phase conformation of Robo1. Three glycoforms of Robo1 from two batches of expressed Robo1 were investigated. The first batch was expressed in FreeStyleTM 293-F cells to produce wild-type Robo1 which exhibits heterogeneous mixture of N-glycans. The second batch was expressed in HEK293S (GnTI-) cells to produce a high-mannose form of Robo1. For comparison purposes, some of these products were treated with EndoF1, which specifically cleaves in between two GlcNAc residues of the core structure of N-glycan to generate the monosaccharide form of Robo1.
The native MS of different glycoforms of Robo1 are shown in Figure 6. Three peaks that correspond to +11 charge state of Robo1 with one GlcNAc attached, Robo1 with a full length, high mannose form of N-glycan attached (Man5GlcNAc2), and Robo1 with heterogeneous, full length N-glycans attached (Gal3GlcNAc3Man3GlcNAc2Fuc1), were selected for the further IM experiments. Regardless of glycoforms, two IM peaks corresponding to two conformations were observed. The compact form of Robo1 was much more abundant for the glycoforms with full-length glycans than for the monosaccharide form. These observations indicated that increasing the size of N-glycan shifted the conformational equilibrium of Robo1 and preferentially stabilized the more compact conformation. Robo1 used to generate the crystal structures did not have any N-glycan attached, which may explain why the more compact conformation was not resolved by X-ray crystallography before [18].
(a) Native ESI mass spectra of different glycoforms of Robo1. (b) Arrival time distributions (ms) of +11 charge state of different glycoforms of Robo1 with and without the binding of a fully sulfated HS dp6. The relative ion population areas of the compact conformation for each glycosylated form of Robo1 are 20.1 ± 0.2, 41.3 ± 0.9, and 40.3 ± 0.8%, respectively. The relative ion population areas of the compact conformation for each glycoform of Robo1-HS dp6 complex are 51.7 ± 1.1, 46.7 ± 0.4, and 45.3 ± 0.9%, respectively
We then incubated Robo1 of different glycoforms with a fully sulfated HS dp6 and measured the ion mobility profiles of formed complexes. As shown in Figure 6b, the compact conformation of Robo1in complexed form is still the dominant peak regardless of the attached N-glycan, indicating that HS still maintained its preference for the compact conformation of Robo1, and this preference was not dramatically affected by altering the size and composition of the attached N-glycan.
Altogether, we observed that Robo1 stayed in two conformations regardless of the variations of the size and composition of the N-glycan component of Robo1, but the relative abundance of each conformation varied. Though a full-length N-glycan had a higher stabilizing effect for the more compact conformation, it did not have a substantial effect on the binding preference of HS towards the compact form of Robo1.
Gas-Phase Stability of Robo1 and Robo1-HS Complexes
The impacts of HS binding on the stability of the three-dimensional structures of Robo1 and Robo1 complexes were examined by collision-induced unfolding (CIU) experiments. In these experiments, trapping collisional energy (CE) were applied to selected Robo1 and Robo1-HS complex ions to gradually increase their internal energy, while the resulting conformational transition from a fully folded state to a fully unfolded state due to collision activation was tracked. The resulting CIU fingerprints of the 10+ charge state of Robo1 and Robo1-HS complexes are shown in Figure 7 as contour plots, in which the drift time and intensity of each feature that emerged during the unfolding process was plotted against the trap CE, which was ramped from 0 to 80 V in 2.5 V increments.
The 2D–contour CIU fingerprints of unbound Robo1 and Robo1-HS complexes, with HS of increasing size from dp4 to dp12. The ion intensities are indicated by a color axis. As the size of HS increases, the conformational stability of the complex increases accordingly, but HS longer than dp8 prevented the complete unfolding of Robo1 upon activation
The CIU fingerprint of unbound Robo1 exhibits three features. The first feature, which has the lowest drift time, corresponds to a natively folded, compact conformation. A second feature appeared at a CE of 10 V and stayed stabilized up to 30 V, corresponding to an intermediate state or a more extended, partially unfolded conformation. As the CE was increased to 80 V, the third feature started to emerge, indicating that Robo1 has completely unfolded.
The CIU fingerprints of 1:1 Robo1-HS dp4/dp6 complexes showed similar unfolding behavior. Compared with unbound Robo1, the most compact and the partially unfolded features showed a more elongated shape while the fully unfolded feature evolved at much higher voltages, indicating that all three features were dramatically stabilized upon the binding of these short HS oligosaccharides. In contrast to the complexes formed with HS dp4/dp6, the CIU fingerprints of Robo1-HS dp8/dp10/dp12 complexes exhibited only two principal conformational features which evolved at similar CE. The absence of a fully unfolded feature indicated that interacting with HS oligosaccharide larger than dp8 induced higher stability against collision-induced activation and protected the protein from completely unfolding. To track the degree of stabilization due to binding of HS, the collision energy required for the transition of the most compact thus the most native feature to a more extended state for Robo1 and different Robo1-HS complexes have been summarized in Supplementary Table 1. In our previous study on the less specific FGF1-HS interactions, we observed that the degree of gas-phase stabilization of FGF1-HS complexes directly correlated with the size of the HS: the larger the HS oligosaccharide, the higher the gas-phase stability of HS bound complexes. In contrast, for the current case of Robo1-HS interaction, we observed comparable unfolding features and comparable transitional CE for the most native feature for the 10+ charge state of Robo1-dp8/dp10/dp12 complexes, indicating that there was no dramatic increase of gas-phase stability as the size of HS becomes larger than dp8. The CIU features of the 9+ charge state of Robo1 and selected Robo1-HS complexes were also studied and their CIU fingerprints are shown in Supplementary figure 4. Regardless of the charge state of choice, the overall trend of increased stability for Robo1 complexes due to the binding of HS remained the same, and there was no dramatic increase of gas-phase stability as the size of HS becomes larger than dp8. Taken together with our observations that HS dp8 has the highest HS binding affinity in the competition experiment and adding extra saccharide units failed to improve the binding affinity, collective evidence supports the hypothesis that the binding pocket/motif on Robo1 is about the size of a dp8, and no larger HS oligosaccharide is required for a high affinity interaction with Robo1 to stabilize Robo1 against unfolding.
Conclusions
This study focused on the application of TWIMS to investigate the conformational details of the interactions between Robo1 and a series of HS oligosaccharides of different primary sequence and size. Robo1 exhibits two interchangeable conformations in the gas phase, one with a cross section that matches the structure previously reported using X-ray crystallography and a previously unreported, more compact structure which exhibits a higher binding affinity toward HS. The data suggests that HS mediates the assembly of Robo1-Slit-HS complexes based on stabilizing or promoting Robo1-Slit binding through a conformational change mechanism. Studies using different glycoforms of Robo1 suggest that this mechanism is independent of the composition and size of N-glycan attached to Robo1.
The results also suggest that the Robo1-HS interaction specifically recognizes a structural motif on HS. Alternating the pattern of sulfation on the sequence had a measurable effect on the degree of binding. Though the actual sequence cannot be defined yet, the observations suggest that this structural motif is about the size of a dp8 and is preferentially modified with N- and 6-O-sulfation of GlcN, and weakened by 2-O sulfation of IdoA. Not only did this structural motif mediate the conformation change involved in Robo1-HS interaction, it also showed the highest binding affinity toward Robo1 and was capable of protecting Robo1 from collision-activated unfolding. Collectively, these observations provide new insights into the way HS mediates Robo1-Slit interactions. These results also highlighted the potential of using heparin/HS-based agents to regulate biological processes mediated by Robo1-Slit interactions.
References
Kennedy, T.E., Serafini, T., de la Torre, J., Tessier-Lavigne, M.: Netrins are diffusible chemotropic factors for commissural axons in the embryonic spinal cord. Cell. 78, 425–435 (1994)
Long, H., Sabatier, C., Ma, L., Plump, A., Yuan, W., Ornitz, D.M., Tamada, A., Murakami, F., Goodman, C.S., Tessier-Lavigne, M.: Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron. 42, 213–223 (2004)
Lartillot, N., Philippe, H.: Improvement of molecular phylogenetic inference and the phylogeny of Bilateria. Phil. Trans. Royal Soc. London B: Biol. Sci. 363, 1463–1472 (2008)
Borrell, V., Cárdenas, A., Ciceri, G., Galcerán, J., Flames, N., Pla, R., Nóbrega-Pereira, S., García-Frigola, C., Peregrín, S., Zhao, Z.: Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron. 76, 338–352 (2012)
Englund, C., Steneberg, P., Falileeva, L., Xylourgidis, N., Samakovlis, C.: Attractive and repulsive functions of Slit are mediated by different receptors in the Drosophila trachea. Development. 129, 4941–4951 (2002)
Andrews, W., Barber, M., Hernadez-Miranda, L.R., Xian, J., Rakic, S., Sundaresan, V., Rabbitts, T.H., Pannell, R., Rabbitts, P., Thompson, H.: The role of Slit-Robo signaling in the generation, migration and morphological differentiation of cortical interneurons. Dev. Biol. 313, 648–658 (2008)
Di Meglio, T., Nguyen-Ba-Charvet, K.T., Tessier-Lavigne, M., Sotelo, C., Chédotal, A.: Molecular mechanisms controlling midline crossing by precerebellar neurons. J. Neurosci. 28, 6285–6294 (2008)
Rama, N., Dubrac, A., Mathivet, T., Chárthaigh, R.-A.N., Genet, G., Cristofaro, B., Pibouin-Fragner, L., Ma, L., Eichmann, A., Chédotal, A.: Slit2 signaling through Robo1 and Robo2 is required for retinal neovascularization. Nat. Med. 21, 483–491 (2015)
Whitford, K.L., Marillat, V., Stein, E., Goodman, C.S., Tessier-Lavigne, M., Chédotal, A., Ghosh, A.: Regulation of cortical dendrite development by Slit-Robo interactions. Neuron. 33, 47–61 (2002)
Wang, L.J., Zhao, Y., Han, B., Ma, Y.G., Zhang, J., Yang, D.M., Mao, J.W., Tang, F.T., Li, W.D., Yang, Y.: Targeting Slit–Roundabout signaling inhibits tumor angiogenesis in chemical-induced squamous cell carcinogenesis. Canc. Sci. 99, 510–517 (2008)
Tseng, R.-C., Lee, S.-H., Hsu, H.-S., Chen, B.-H., Tsai, W.-C., Tzao, C., Wang, Y.-C.: SLIT2 attenuation during lung cancer progression deregulates β-catenin and E-cadherin and associates with poor prognosis. Canc. Res. 70, 543–551 (2010)
Schmid, B.C., Rezniczek, G.A., Fabjani, G., Yoneda, T., Leodolter, S., Zeillinger, R.: The neuronal guidance cue Slit2 induces targeted migration and may play a role in brain metastasis of breast cancer cells. Breast Canc. Res. Treat. 106, 333–342 (2007)
Wang, B., Xiao, Y., Ding, B.-B., Zhang, N., Yuan, X.-B., Gui, L., Qian, K.-X., Duan, S., Chen, Z., Rao, Y.: Induction of tumor angiogenesis by Slit-Robo signaling and inhibition of cancer growth by blocking Robo activity. Canc. Cell. 4, 19–29 (2003)
Xian, J., Clark, K.J., Fordham, R., Pannell, R., Rabbitts, T.H., Rabbitts, P.H.: Inadequate lung development and bronchial hyperplasia in mice with a targeted deletion in the Dutt1/Robo1 gene. Proc. Natl. Acad. Sci. 98, 15062–15066 (2001)
Dallol, A., Krex, D., Hesson, L., Eng, C., Maher, E.R., Latif, F.: Frequent epigenetic inactivation of the SLIT2 gene in gliomas. Oncogene. 22, 4611–4616 (2003)
Walmod, P. S., Pedersen, M. V., Berezin, V. , Bock, E.: Cell adhesion molecules of the immunoglobulin superfamily in the nervous system; In Handbook of neurochemistry and molecular neurobiology. Springer 35-151 (2007)
Evans, T.A., Bashaw, G.J.: Functional diversity of Robo receptor immunoglobulin domains promotes distinct axon guidance decisions. Curr. Biol. 20, 567–572 (2010)
Morlot, C., Thielens, N.M., Ravelli, R.B., Hemrika, W., Romijn, R.A., Gros, P., Cusack, S., McCarthy, A.A.: Structural insights into the Slit-Robo complex. Proc. Natl. Acad. Sci. 104, 14923–14928 (2007)
Fukuhara, N., Howitt, J.A., Hussain, S.-A., Hohenester, E.: Structural and functional analysis of slit and heparin binding to immunoglobulin-like domains 1 and 2 of Drosophila Robo. J. Biol. Chem. 283, 16226–16234 (2008)
Lindahl, U., Li, J.: p., Interactions between heparan sulfate and proteins—design and functional implications. Int. Rev. Cell Molec. Biol. 276, 105–159 (2009)
Xu, D., Esko, J.D.: Demystifying heparan sulfate–protein interactions. Ann. Rev. Biochem. 83, 129–157 (2014)
Bülow, H.E., Hobert, O.: Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron. 41, 723–736 (2004)
Johnson, K.G., Ghose, A., Epstein, E., Lincecum, J., O'Connor, M.B., Van Vactor, D.: Axonal heparan sulfate proteoglycans regulate the distribution and efficiency of the repellent slit during midline axon guidance. Curr. Biol. 14, 499–504 (2004)
Steigemann, P., Molitor, A., Fellert, S., Jäckle, H., Vorbrüggen, G.: Heparan sulfate proteoglycan syndecan promotes axonal and myotube guidance by slit/robo signaling. Curr. Biol. 14, 225–230 (2004)
Hu, H.: Cell-surface heparan sulfate is involved in the repulsive guidance activities of Slit2 protein. Nat. Neurosci. 4, 695–701 (2001)
Hussain, S.-A., Piper, M., Fukuhara, N., Strochlic, L., Cho, G., Howitt, J.A., Ahmed, Y., Powell, A.K., Turnbull, J.E., Holt, C.E.: A molecular mechanism for the heparan sulfate dependence of slit-robo signaling. J. Biol. Chem. 281, 39693–39698 (2006)
Ypsilanti, A. R. , Chedotal, A.: Roundabout receptors; In Cell Adhesion Molecules. Springer 133-164 (2014)
Hou, S.T., Jiang, S.X., Smith, R.A.: Permissive and repulsive cues and signalling pathways of axonal outgrowth and regeneration. Int. Rev. Cell Molec. Biol. 267, 125–181 (2008)
Ballard, M.S., Hinck, L.: A roundabout way to cancer. Adv. Canc. Res. 114, 187 (2012)
Zhao, Y., Singh, A., Li, L., Linhardt, R.J., Xu, Y., Liu, J., Woods, R.J., Amster, I.J.: Investigating changes in the gas-phase conformation of Antithrombin III upon binding of Arixtra using traveling wave ion mobility spectrometry (TWIMS). Analyst. 140, 6980–6989 (2015)
Zhao, Y., Singh, A., Xu, Y., Zong, C., Zhang, F., Boons, G.-J., Liu, J., Linhardt, R.J., Woods, R.J., Amster, I.J.: Gas-Phase Analysis of the Complex of Fibroblast GrowthFactor 1 with Heparan Sulfate: A Traveling Wave Ion Mobility Spectrometry (TWIMS) and Molecular Modeling Study. J. Am. Soc. Mass Spectrom. 28, 96–109 (2017)
Liu, Z., Patel, K., Schmidt, H., Andrews, W., Pini, A., Sundaresan, V.: Extracellular Ig domains 1 and 2 of Robo are important for ligand (Slit) binding. Molec. Cell. Neurosci. 26, 232–240 (2004)
Dickson, B.J., Gilestro, G.F.: Regulation of commissural axon pathfinding by slit and its Robo receptors. Annu. Rev. Cell Dev. Biol. 22, 651–675 (2006)
Xu, Y., Cai, C., Chandarajoti, K., Hsieh, P.-H., Li, L., Pham, T.Q., Sparkenbaugh, E.M., Sheng, J., Key, N.S., Pawlinski, R.: Homogeneous low-molecular-weight heparins with reversible anticoagulant activity. Nat. Chem. Biol. 10, 248–250 (2014)
Zong, C., Venot, A., Dhamale, O., Boons, G.-J.: Fluorous supported modular synthesis of heparan sulfate oligosaccharides. Org. Lett. 15, 342–345 (2013)
Zhang, F., Moniz, H.A., Walcott, B., Moremen, K.W., Wang, L., Linhardt, R.J.: Probing the impact of GFP tagging on Robo1-heparin interaction. Glycoconjug. J. 31, 299–307 (2014)
Beckett, D., Kovaleva, E., Schatz, P.J.: A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929 (1999)
Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T.C., Waldo, G.S.: Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006)
Carrington, J.C., Dougherty, W.G.: A viral cleavage site cassette: identification of amino acid sequences required for tobacco etch virus polyprotein processing. Proc. Natl. Acad. Sci. 85, 3391–3395 (1988)
Reeves, P.J., Callewaert, N., Contreras, R., Khorana, H.G.: Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. 99, 13419–13424 (2002)
Meng, L., Forouhar, F., Thieker, D., Gao, Z., Ramiah, A., Moniz, H., Xiang, Y., Seetharaman, J., Milaninia, S., Su, M.: Enzymatic basis for N-Glycan Sialylation STRUCTURE OF RAT α2, 6-SIALYLTRANSFERASE (ST6GAL1) REVEALS CONSERVED AND UNIQUE FEATURES FOR GLYCAN SIALYLATION. J. Biol. Chem. 288, 34680–34698 (2013)
Eschweiler, J.D., Rabuck-Gibbons, J.N., Tian, Y., Ruotolo, B.T.: CIUSuite: a quantitative analysis package for collision induced unfolding measurements of gas-phase protein ions. Anal. Chem. 87, 11516–11522 (2015)
Ruotolo, B.T., Benesch, J.L., Sandercock, A.M., Hyung, S.-J., Robinson, C.V.: Ion mobility–mass spectrometry analysis of large protein complexes. Nat. Protoc. 3, 1139–1152 (2008)
Gao, Q., Chen, C.-Y., Zong, C., Wang, S., Ramiah, A., Prabhakar, P., Morris, L.C., Boons, G.-J., Moremen, K.W., Prestegard, J.H.: Structural Aspects of Heparan Sulfate Binding to Robo1–Ig1–2. ACS Chem. Biol. 11, 3106–3113 (2016)
Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C., Ferrin, T.E.: UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Fiser, A., Šali, A.: Modeller: generation and refinement of homology-based protein structure models. Meth. Enzymol. 374, 461–491 (2003)
Hornak, V., Abel, R., Okur, A., Strockbine, B., Roitberg, A., Simmerling, C.: Comparison of multiple Amber force fields and development of improved protein backbone parameters. Prot.: Struct. Funct. Bioinform. 65, 712–725 (2006)
Kirschner, K.N., Yongye, A.B., Tschampel, S.M., González-Outeiriño, J., Daniels, C.R., Foley, B.L., Woods, R.J.: GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 29, 622–655 (2008)
Salomon-Ferrer, R., Götz, A.W., Poole, D., Le Grand, S., Walker, R.C.: Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theor. Comput. 9, 3878–3888 (2013)
Case, D.A., Cheatham, T.E., Darden, T., Gohlke, H., Luo, R., Merz, K.M., Onufriev, A., Simmerling, C., Wang, B., Woods, R.J.: The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005)
Darden, T., York, D., Pedersen, L.: Particle mesh Ewald: An N· log (N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993)
Roe, D.R., Cheatham III, T.E.: PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 9, 3084–3095 (2013)
Humphrey, W., Dalke, A., Schulten, K.: VMD: visual molecular dynamics. J. Molec. Graph. 14, 33–38 (1996)
Varki, A., Cummings, R.D., Aebi, M., Packer, N.H., Seeberger, P.H., Esko, J.D., Stanley, P., Hart, G., Darvill, A., Kinoshita, T.: Symbol nomenclature for graphical representations of glycans. Glycobiology. 25, 1323–1324 (2015)
Thieker, D.F., Hadden, J.A., Schulten, K., Woods, R.J.: 3D implementation of the symbol nomenclature for graphical representation of glycans. Glycobiology. 26, 786–787 (2016)
Zhang, F., Moniz, H.A., Walcott, B., Moremen, K.W., Linhardt, R.J., Wang, L.: Characterization of the interaction between Robo1 and heparin and other glycosaminoglycans. Biochimie. 95, 2345–2353 (2013)
Irie, A., Yates, E.A., Turnbull, J.E., Holt, C.E.: Specific heparan sulfate structures involved in retinal axon targeting. Development. 129, 61–70 (2002)
Zong, C., Huang, R., Condac, E., Chiu, Y., Xiao, W., Li, X., Lu, W., Ishihara, M., Wang, S., Ramiah, A.: Integrated Approach to Identify Heparan Sulfate Ligand Requirements of Robo1. J. Am. Chem. Soc. 138, 13059–13067 (2016)
Li, Z., Moniz, H., Wang, S., Ramiah, A., Zhang, F., Moremen, K.W., Linhardt, R.J., Sharp, J.S.: High structural resolution hydroxyl radical protein footprinting reveals an extended Robo1-heparin binding interface. J. Biol. Chem. 290, 10729–10740 (2015)
Jurneczko, E., Barran, P.E.: How useful is ion mobility mass spectrometry for structural biology? The relationship between protein crystal structures and their collision cross sections in the gas phase. Analyst. 136, 20–28 (2011)
Benesch, J.L., Ruotolo, B.T.: Mass spectrometry: come of age for structural and dynamical biology. Curr. Opin. Struct. Biol. 21, 641–649 (2011)
Johnson, K.G., Van Vactor, D.: Receptor protein tyrosine phosphatases in nervous system development. Physiol. Rev. 83, 1–24 (2003)
Coles, C.H., Shen, Y., Tenney, A.P., Siebold, C., Sutton, G.C., Lu, W., Gallagher, J.T., Jones, E.Y., Flanagan, J.G., Aricescu, A.R.: Proteoglycan-specific molecular switch for RPTPσ clustering and neuronal extension. Science. 332, 484–488 (2011)
Reichert, M.C., Brown, H.E., Evans, T.A.: In vivo functional analysis of Drosophila Robo1 immunoglobulin-like domains. Neural Dev. 11, 15 (2016)
Seo, Y., Andaya, A., Bleiholder, C., Leary, J.A.: Differentiation of CC vs CXC chemokine dimers with GAG octasaccharide binding partners: an ion mobility mass spectrometry approach. J. Am. Chem. Soc. 135, 4325–4332 (2013)
Acknowledgements
The authors are grateful for generous support from the National Institutes of Health (P41GM103390). R.J.W. thanks the National Institutes of Health for grant support (U01 CA207824).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 680 kb)
Rights and permissions
About this article
Cite this article
Zhao, Y., Yang, J.Y., Thieker, D.F. et al. A Traveling Wave Ion Mobility Spectrometry (TWIMS) Study of the Robo1-Heparan Sulfate Interaction. J. Am. Soc. Mass Spectrom. 29, 1153–1165 (2018). https://doi.org/10.1007/s13361-018-1903-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-018-1903-4