Abstract
Background
The p97/valosin-containing protein (VCP) complex is a crucial factor for the segregation of ubiquitinated proteins in the DNA damage response and repair pathway.
Objective
We investigated whether blocking the p97/VCP function can inhibit the proliferation of RepID-deficient cancer cells using immunofluorescence, clonogenic survival assay, fluorescence-activated cell sorting, and immunoblotting.
Result
p97/VCP was recruited to chromatin and colocalized with DNA double-strand breaks in RepID-deficient cancer cells that undergo spontaneous DNA damage. Inhibition of p97/VCP induced death of RepID-depleted cancer cells. This study highlights the potential of targeting p97/VCP complex as an anticancer therapeutic approach.
Conclusion
Our results show that RepID is required to prevent excessive DNA damage at the endogenous levels. Localization of p97/VCP to DSB sites was induced based on spontaneous DNA damage in RepID-depleted cancer cells. Anticancer drugs targeting p97/VCP may be highly potent in RepID-deficient cells. Therefore, we suggest that p97/VCP inhibitors synergize with RepID depletion to kill cancer cells.
Similar content being viewed by others
Introduction
DNA double-strand break (DSB) is a form of damage, wherein the phosphate backbones of the two complementary DNA strands are simultaneously broken, leading to serious consequences including chromosomal aberrations or cell death (Mehta and Haber 2014). DSBs may be caused by exogenous factors, such as chemical mutagens, chemotherapeutic DNA damaging agents, ionizing radiation, or by endogenous stressors including abnormal DNA replication and reactive oxygen species (Chen and Stubbe 2005, Wyrobek et al. 2005). During replication, prolonged stalling of replication forks results in the generation of extended stretches of single-stranded DNA (ssDNA). Persistent replication stress causes fork collapse, resulting in highly lethal DNA DSBs (Lopes et al. 2001; Aladjem and Redon 2017; Kurat et al. 2017). A cascade of protein modifications involving relocalization and degradation occurs in response to DSBs as follows: the Mre11-rad50-Ns1 (MRN) complex (sensor) recruits Ataxia telangiectasia mutated (ATM) kinase, which is known as the master factor at the DSB sites (Fishman-Lobell, Rudin et al. 1992; Manis, Morales et al. 2004; Ma, Lu et al. 2005; Difilippantonio, Gapud et al. 2008; Bunting, Callen et al. 2012), followed by rapid phosphorylation of H2AX at serine 139 in an ATM-dependent manner (Burma, Chen et al. 2001; Fernandez-Capetillo, Lee et al. 2004). Amplification of this signal can lead to the subsequent recruitment of several proteins, including E3 ubiquitin ligase ring finger protein 8 (RNF8) and RNF168. This ubiquitination-based decision of localization or degradation of the downstream factors facilitates DSB repair by homologous recombination (HR) and nonhomologous end joining (NHEJ) pathways (Huyen, Zgheib et al. 2004; Doil, Mailand et al. 2009; Stewart, Panier et al. 2009; Symington and Gautier 2011; Mallette, Mattiroli et al. 2012).
The AAA + (Associated with diverse cellular Activities) ATPase p97, also known as valosin-containing protein (p97/VCP), is a central component of the ubiquitin–proteasome system (Dantuma and Hoppe 2012; Vaz, Halder et al. 2013; Meyer and Weihl 2014). P97/VCP segregates ubiquitinated substrates involved in DNA replication, transcription, repair, and cell cycle progression using ATP-derived energy for degradation or recycling (Torrecilla, Oehler et al. 2017). DNA repair proteins breast cancer type 1 (BRCA1) and Werner helicase interact with p97/VCP (Zhang, Wang et al. 2000; Partridge, Lopreiato et al. 2003; Indig, Partridge et al. 2004), which physically associates with DSBs and removes ubiquitinated lethal 3 malignant brain tumor 1 (L3MBTL1), Ku, and KRAB-associated protein 1 (KAP1) for the subsequent loading of 53BP1, BRCA1, and Rad51 (Acs, Luijsterburg et al. 2011, Meerang, Ritz et al. 2011, Kuo, Li et al. 2016, van den Boom, Wolf et al. 2016). Inactivation or depletion of the p97/VCP-mediated segregation system compromises HR and NHEJ pathways, demonstrating the essential role of p97/VCP in DNA repair (Acs, Luijsterburg et al. 2011; Meerang, Ritz et al. 2011).
The replication origin-binding protein RepID, also known as Pleckstrin homology domain-interacting protein (PHIP) and DDB1/CUL4-associated factor 14 (DCAF14), is a multifunctional protein that facilitate proliferation of cancer cells (Farhang-Fallah, Randhawa et al. 2002; Podcheko, Northcott et al. 2007; Tokuda, Fujita et al. 2007; Li, Francisco et al. 2010; De Semir, Nosrati et al. 2012; de Semir, Bezrookove et al. 2018). Chromatin-localized RepID selectively associates with a subgroup of replication origin proteins to initiate DNA replication, possibly by regulating the formation of chromatin loops (Zhang, Huang et al. 2016). RepID-depleted cells show reduced initiation frequency as well as slow elongation and frequent stalling at replication forks (Zhang, Huang et al. 2016). The Cullin-RING ubiquitin ligase complex 4 (CRL4) is recruited to the chromatin by RepID (Jang, Redon et al. 2018a, b; Jang, Zhang et al. 2018a, b). In the absence of RepID, CRL4 cannot ubiquitinate substrates localized on the chromatin, resulting in re-replication via accumulation of the DNA replication origin licensing factor CDT1 or delay in the transition from metaphase to anaphase during mitosis owing to the failure to degrade BUB3, a subunit of the spindle assembly checkpoint proteins blocking the anaphase-promoting complex/cyclosome ubiquitin E3 ligase (APC/C)(Jang, Nathans et al. 2020a, b; Jang, Redon et al. 2020a, b). These abnormal replication and chromosomal segregation patterns in the absence of RepID suggest that RepID plays a role in the prevention of DNA damage during cell proliferation.
Here, we report that RepID is a safeguard protein that prevents excessive endogenous DNA damage. We observed that RepID-depleted cells showed increased levels of DSBs. This event correlated with higher recruitment of p97/VCP on the chromatin in the absence of RepID, leading to hypersensitivity of cancer cells to the p97/VCP-targeting inhibitor CB5083. Our findings suggest that RepID levels could be investigated as possible effectors of cancer therapy based on the DNA damage-p97/VCP axis.
Materials and methods
Cell culture and chemicals
Human U2OS osteosarcoma cells with and without RepID were incubated in Dulbecco’s modified Eagle’s medium (Invitrogen, 10,569–010) supplemented with 10% heat-inactivated fetal bovine serum in a 37 °C/5% CO2 humidified incubator. U2OS original cancer cell lines were obtained from The American Type Culture Collection (ATCC; www.atcc.org). All cell lines were tested negative for mycoplasmas (Lonza, LT07-418). P97/VCP inhibitor CB5083 (Selleckchem, S8101) or CPT (Selleckchem, S1288) was added to the medium at indicated concentrations.
RepID-depleted cell lines
RepID was depleted in U2OS cells using the CRISPR/CAS9 tool. A 20 base-pair guide sequence targeting the fifth exon of RepID (5ʹ-CTGCAAATATGTCATCGACTAGG-3ʹ) and the eighth exon of RepID (5ʹ-GTGATAAAATGATCCGAGTCTGG-3ʹ) in U2OS cells was selected from a published database of predicted high-specificity protospacer-PAM target sites in the human exome. Cells were cultured in six-well dishes to 70%-80% confluency for co-transfection with 2 μg RepID single-guide RNA (sgRNA) plasmid, 2 μg linearized pCR2.1 vector harboring a puromycin-resistance gene expression cassette, and 10 μL Lipofectamine 2000 (Life Technologies). Cloning, selection, and verification were performed using polymerase chain reaction (PCR).
Fluorescence-activated cell sorting (FACS) analysis
Cells were labeled with 10 μM EdU for 30 min before harvesting using the Click-iT EdU kit (Invitrogen, C10424). Cell staining was performed according to the manufacturer’s protocol. We used 4′,6-diamidino-2-phenylindole (DAPI) for DNA counterstaining. An LSR Fortessa cell analyzer (BD Biosciences) with FlowJo 10.5.2 software was used for cell cycle analyses. All experiments reported representative results of at least three independent repetitions.
Clonogenic survival assay
U2OS cells were plated in six-well plates (500 cells/well) in triplicates and treated with CB5083 for 10 days. Colonies were fixed with 1% paraformaldehyde (PFA) and stained with crystal violet. Well intensity was measured using ImageJ software. All experiments reported representative results of at least three independent repetitions.
Immunofluorescence analysis
U2OS cells were incubated in phosphate-buffered saline (PBS)-T (0.2% Triton X-100 in 1 × PBS, protease inhibitor cocktail [Sigma, P8340], phenylmethylsulfonyl fluoride [PMSF], and phosphatase inhibitor cocktail [Roche, P4906845001]) for 5 min on ice, and then fixed with 2% PFA. Primary antibody staining was performed as follows: anti-phosphorylated ATM (Thermo Fisher Scientific, MA5-15,185, 1:200), anti-53BP1 (Novus, NB100-305A, 1:200), anti-p97/VCP (Abcam, ab11433, 1:100), and anti-γH2AX (Millipore, 05–636, 1:1000) for 3 h at room temperature. Secondary antibody staining was performed as follows: Alexa 488-conjugated anti-mouse IgG and Alexa 555-conjugated anti-rabbit IgG (1:500, Thermo Fisher Scientific, A11029 and A21428). EdU was detected using the Click-iT assay kit according to the manufacturer’s protocol. A Zeiss LSM710 confocal microscope and a Visitech VT-ISIM were used, and coefficients were generated using ImageJ software and plugins.
Chromatin fractionation and immunoblotting
Harvested U2OS cells were incubated in a cytosol extraction buffer containing NP-40 (20 mM Tris–HCl pH 7.4, 10 mM sodium chloride [NaCl], 3 mM magnesium chloride [MgCl2], 0.5% NP-40, PMSF, protease inhibitor cocktail, and phosphatase inhibitor cocktail). Cells were harvested by centrifugation at 3000×g for 5 min at 4 °C, washed, and resuspended in a nuclear extraction buffer (10 mM Tris–HCl pH 7.4, 100 mM NaCl, 1% Triton X-100, 1 mM ethylenediaminetetraacetic acid [EDTA] pH 8, 1 mM ethylene glycol tetraacetic acid [EGTA], 0.1% sodium dodecyl sulfate [SDS], 10% glycerol, 0.5% sodium deoxycholate, protease inhibitor cocktail, and phosphatase inhibitor cocktail). The suspension was vortexed, incubated on ice, and then centrifuged at 5000 × g for 5 min at 4 °C. The pellet was resuspended in nuclear extraction buffer containing 5 mM calcium chloride (CaCl2) and micrococcal nuclease (New England Biolabs, Cat. M0247S), vortexed, and incubated at 37 °C for 5 min. Chromatin-bound fractions were collected after centrifugation at 18,000×g for 5 min at 4 °C. Total cell lysates and chromatin-bound proteins were detected by SDS–polyacrylamide gel electrophoresis. The following primary antibodies were used: anti-RepID (NCI186, 1:1,000), anti-p97/VCP (Abcam, ab11433, 1:2,000), anti-γH2AX (Millipore, 05–636, 1:2,000), anti-α-tubulin (Sigma, T9026, 1:2,000), and anti-histone H3 (Millipore, 07–690, 1:20,000). For secondary antibodies, horseradish peroxidase (HRP)-linked anti-mouse IgG (Cell Signaling, 7076) and HRP-linked anti-rabbit IgG (Cell Signaling, 7074) were used following the manufacturer’s protocols.
Results
RepID prevents excessive DNA damage
To determine whether RepID plays a role in regulating the generation of DSBs, we first measured the levels of phosphorylated ATM, the master regulator of DNA damage response in U2OS human osteosarcoma cell lines, which were deficient of RepID (RepID knock-out [KO]). RepID depletion was performed using the clustered regularly interspaced short palindromic repeats (CRISPR)/caspase 9 (CAS9) gene editing tool and single clone was selected followed by verification (Jang, Zhang et al. 2018a, b). Confocal microscopy showed that phosphorylated ATM foci were more frequent in RepID-deficient cells than in RepID-proficient cells; this phenotype was consistent during all cell cycles, including EdU-positive S-phase or EdU-negative G1/G2 phase (Fig. 1a, c, dimethyl sulfoxide [DMSO] panel). Treatment with camptothecin (CPT), a DSB-inducing agent that inhibits topoisomerase I (Wall and Wani 1995), significantly induced phosphorylated ATM signals that were stronger in RepID-deficient cells than in RepID-proficient cells (Fig. 1a, c, CPT panel). Consistent with the observed behavior of ATM foci, foci with the DSB marker γH2AX and the NHEJ player 53BP1 were notably detected after RepID depletion (Fig. 1b, d, e). The colocalization of 53BP1 and γH2AX increased in CPT-treated RepID KO cells (Fig. 1f). These results suggest that RepID prevents excessive DNA damage.
RepID prevents excessive DNA damage during cell cycle progression. a, b RepID proficient (Wild Type—WT) or RepID-depleted (Knock Out—KO) U2OS cells were incubated with/without 0.5 M CPT for 1 h. EdU-labeled, pre-extracted, and phosphorylated ATM (green) (a), γH2AX (green), and 53BP1 (red) (b), DNA content (DAPI) and EdU (magenta) were detected. Cells in replicating S-phase (EdU-positive) or non-replicating G1/G2 phase (EdU-negative) were identified by EdU staining. Scale bar indicates 10 m. (c-e) Relative intensity. Phosphorylated ATM (c), 53BP1 (d), and γH2AX (E). (f) Extent of colocalization between 53BP1 and γH2AX. Pearson’s correlation coefficients (n = 20). p-values were calculated using a two-tailed t-test, and error bars represent standard deviation from three independent experiments (***p-value < 0.001)
Recruitment of P97/VCP to DSB sites increases RepID-deficient cancer cells
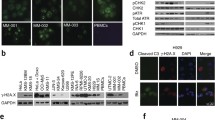
P97/VCP segregase is an essential factor regulating DSB repair by orchestrating the segregation of ubiquitinated DSB repair proteins (Torrecilla, Oehler et al. 2017). We investigated p97/VCP localization in the RepID-deficient cells which generate excessive DSBs. Nuclei were pre-extracted to remove soluble nuclear proteins and incubated with p97/VCP and γH2AX antibodies. Most p97/VCP was detected in the nucleoli of RepID-proficient and -deficient cells (Fig. 2a, DMSO panel). However, in RepID-deficient cells, some p97/VCP was detected outside the nucleoli and colocalized with γH2AX foci (Fig. 2a, b, DMSO panel). RepID-proficient cells also showed some colocalization of p97/VCP and γH2AX after CPT treatment along with an increase in γH2AX foci together with diffused patterns of p97/VCP (Fig. 2a, b, CPT panel). However, intensity profile analyses revealed that the colocalization of p97/VCP and γH2AX was more evident in CPT-treated RepID-deficient cells (Fig. 2a, b, CPT panel) than in RepID-proficient cells.
Recruitment of p97/VCP to DSB sites is increased in RepID-deficient cancer cells. a U2OS cells were treated with/without 1.0 M CPT for 1 h, pre-extracted, and incubated with p97/VCP (green) and γH2AX (red) antibodies together with DAPI (blue). Scale bar indicates 10 m. b Colocalization of p97/VCP and γH2AX was analyzed by intensity profiling as indicated in (a). Gray backgrounds indicate colocalized region. c Levels of p97/VCP from RepID WT or RepID KO U2OS cancer cell lines after exposure to MG132 proteasome inhibitor. Histone H3 and α-tubulin were used as loading controls. Total, total cell lysates; chromatin, chromatin-bound proteins. d Quantification of chromatin-bound p97/VCP normalized to the levels in untreated RepID WT cells. Error bars represent standard deviations from three independent experiments. p-values were calculated using a two-tailed t-test, and error bars represent standard deviation from three independent experiments (***p-value < 0.001)
To evaluate the extent of p97/VCP recruitment to DSBs, we used MG132 that accumulates ubiquitinated proteins, including DSB repair factors, by preventing proteasomal degradation. In the absence of RepID or MG132, we did not observe any effect on the expression of p97/VCP (Fig. 2c). However, the chromatin-bound fractions of p97/VCP significantly increased in RepID-deficient cells (2.43-fold). The recruitment of p97/VCP on the chromatin was notably clearer after MG132 treatment in RepID-depleted cells (6.21-fold in RepID wild-type [WT] vs 13.7-fold in RepID KO cells) (Fig. 2c, d). These results show that p97/VCP recruitment to the chromatin is enhanced along with excessive DNA damage after RepID depletion.
RepID-deficient cancer cells are sensitive to p97/VCP inhibitor
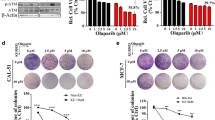
The above observations suggest that the function of p97/VCP might be crucial for DNA repair in RepID-deficient cancer cells. To determine whether p97/VCP-mediated segregation of ubiquitinated proteins during DNA damage response and repair is critical for the proliferation of RepID-deficient cells, both RepID WT and RepID KO cells were exposed to the p97/VCP inhibitor (p97i) CB5083 (Anderson, Le Moigne et al. 2015). As shown in Fig. 3a and b, RepID-deficient cells were more sensitive to p97/VCP inhibition than RepID proficient cells (no surviving colonies in RepID deficient cells treated with 0.6 M p97i). On the other hand, the cells expressing intact RepID were resistant to p97i treatment. A fraction of both RepID-expressing and RepID-depleted cells (SubG1 fraction) died following exposure to CB5083 for up to 4 days. The number of cells in SubG1 was higher in RepID-depleted group than in RepID-expressing group (24 h: 1.36% and 3.61% in WT and KO, respectively; 48 h: 2.28% and 7.06% in WT and KO, respectively; 96 h: 1.03% and 10.3% in WT and KO, respectively) (Fig. 3c, d). Levels of p97/VCP on the chromatin were clearly higher in RepID-depleted cells than in RepID WT cells, consistent with the induced expression of γH2AX following CB5083 treatment (Fig. 3e). Taken together, our observations suggest that RepID expression may modulate the sensitivity of cancer cells to p97/VCP-targeting drugs.
RepID-deficient cancer cells are sensitive to p97/VCP inhibitor. a, b Colony formation assay with U2OS RepID WT and KO cells upon CB5083 treatment (a). Bar charts indicate relative growing intensity by measuring covered area (b). c CB5083-treated RepID WT and KO U2OS cells were labeled with EdU for 30 min and analyzed by flow cytometry. Percentages of cells in each cell cycle phase are indicated in the flow cytometry plots. d Fold changes in the SubG1 phase were based on the values from non-treated RepID WT cells. P-values were calculated using a two-tailed t-test and error bars in all results represent standard deviation from three independent experiments (**p-value < 0.01, ***p < 0.001). e Immunoblot analysis showing elevated accumulation of p97/VCP on the chromatin of RepID-deficient cells in response to CB5083 treatment. The numbers under the panels represent the intensity ratios for p97/VCP normalized by the intensity of the signal in untreated RepID WT or the intensity ratios for γH2AX normalized by each histone H3 signals from three independent experiments
Discussion
The data presented here demonstrate the crucial role of RepID protein in preventing excessive DNA damage at endogenous levels. Fewer replication initiation events and stalled replication forks with asymmetric progression in RepID-depleting cells may cause single-strand DNA breaks (SSBs) by generating excessive ssDNA through the continuous DNA unwinding in a minichromosome maintenance (MCM) helicase-dependent manner (Smith, Fu et al. 2009; Van, Yan et al. 2010). These replication stresses are susceptible to fork collapse, subsequently leading to DSB formation (Pfeiffer, Goedecke et al. 2000). DSB can also occur in the absence of CRL4 recruitment on the chromatin in RepID-deficient cells. Failure to degrade CDT1, known as an origin-licensing factor and a substrate of CRL4, precedes re-replication mediated by CDT1-based re-licensing in RepID KO cells (Machida, Hamlin et al. 2005; Jang, Zhang et al. 2018a, b). Accumulation of other CRL4 substrates such as the histone methyltransferase SET8 or the CDK inhibitor p21, which is required for proper S-phase progression, prevention of spontaneous DNA damage and activation of the G2/M checkpoint, can also generate DSB (Jorgensen, Elvers et al. 2007; Abbas, Sivaprasad et al. 2008; Huen, Sy et al. 2008; Kim, Starostina et al. 2008; Liu, Lee et al. 2009; Tardat, Brustel et al. 2010). The BUB3-mediated delay in the metaphase-anaphase transition, followed by abnormal chromosomal segregation in RepID-deficient cells are also possibly associated with the occurrence of DNA damage foci (Janssen, van der Burg et al. 2011, Jang, Nathans et al. 2020a, b).
RepID’s functional domains mediate its interaction with chromatin and might underlie its role in regulating the DNA damage response. RepID directly interacts with chromatin using its cryptic Tudor domain and bromodomain (Ji, Dadon et al. 2015; Morgan, Rickels et al. 2017). Many bromodomain-containing proteins have been recently identified as crucial factors promoting genome stability (Kim, Lee et al. 2019). In line with previous reports, we observed elevated DSB markers such as phosphorylated ATM, γH2AX, and 53BP1 foci in RepID-deficient cells regardless of whether those cells were actively engaged in DNA synthesis. These observations suggest that RepID is required to protect cells from excessive DNA damage during all phases of the cell cycle. Future studies are warranted to identify proteins that serve as direct targets of CRL4 and how RepID prevents the generation of endogenous DNA damage.
We observed that a p97/VCP-targeting anticancer drug was highly potent in RepID-deficient cancer cells. This finding is in line with the association of p97/VCP function in DSB and repair (Torrecilla, Oehler et al. 2017). Notably, the chromatin association of p97/VCP increased in RepID-deficient cells, correlated with an elevated prevalence of DNA damage. Taken together with the redistribution and mobilization of nucleolar p97/VCP, along with other repair proteins (e.g. WRN), into the nucleoplasm after exposure to CPT, these observations are consistent with the proposed role of the nucleolus as a depository of repair proteins that are rapidly deployed upon exposure to potentially genotoxic perturbations (Yang, Maiguel et al. 2002; Nalabothula, Indig et al. 2010; Indig, Rybanska et al. 2012; Torrecilla et al. 2017).
We propose the following model for the RepID-p97/VCP axis in DNA damage response and repair (Fig. 4). Depletion of RepID results in endogenous DNA damage caused by abnormal replication, attenuation of CRL4 chromatin recruitment and low HR efficiency. This spontaneous DNA damage facilitates p97/VCP chromatin binding to catalyze the segregation of ubiquitinated proteins during the repair process. As RepID-deficient cells exhibit excessive DNA damage, such cells may depend on p97/VCP activity to a higher extent than RepID-proficient cells. This is consistent with the increased potency of p97/VCP inhibitor in RepID-KO cells.
Model depicting the RepID-p97/VCP axis during DNA damage response and repair. In RepID WT cells, DNA replication in S-phase (bidirectional replication as indicated in green and red arrows with same length) or chromosomal segregation during mitosis progresses normally. RepID possibly contributes to the HR DNA repair pathway. In RepID-depleted (RepID KO) cells, excessive DSBs can be generated at the endogenous levels by following reasons: (1) Abnormal DNA replication that includes fewer replication initiation and asymmetric progression (different length of the green and red arrows) resulting in stalled replication forks; (2) accumulation of CRL4 substrates crucial for the prevention of re-replication, spontaneous DNA damage, or mis-segregation of the chromosome owing to compromised CRL4 recruitment on the chromatin; (3) attenuation of HR activity. Thus, p97/VCP recruitment to DSBs increased in RepID KO cells, leading to increased sensitivity to the p97/VCP inhibitor
Our study has potentially significant clinical implications. The 97/VCP network is now regarded as an attractive anticancer target, and CB5083 is currently being tested in clinical trials (Anderson et al. 2015). Our findings provide an important connection that underlies the DNA damage and repair effects of RepID and p97/VCP in cancer and lay the foundation for new strategies to target p97/VCP based on RepID interactions in cancer.
References
Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A (2008) PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev 22(18):2496–2506
Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, Dantuma NP (2011) The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat Struct Mol Biol 18(12):1345–1350
Aladjem MI, Redon CE (2017) Order from clutter: selective interactions at mammalian replication origins. Nat Rev Genet 18(2):101–116
Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, Madriaga A, Soriano F, Menon MK, Wu ZY, Kampmann M, Chen Y, Weissman JS, Aftab BT, Yakes FM, Shawver L, Zhou HJ, Wustrow D, Rolfe M (2015) Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell 28(5):653–665
Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, Lopez-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, Chen HT, Fernandez-Capetillo O, D’Andrea A, Nussenzweig A (2012) BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell 46(2):125–135
Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ (2001) ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 276(45):42462–42467
Chen J, Stubbe J (2005) Bleomycins: towards better therapeutics. Nat Rev Cancer 5(2):102–112
Dantuma NP, Hoppe T (2012) Growing sphere of influence: Cdc48/p97 orchestrates ubiquitin-dependent extraction from chromatin. Trends Cell Biol 22(9):483–491
de Semir D, Bezrookove V, Nosrati M, Dar AA, Wu C, Shen J, Rieken C, Venkatasubramanian M, Miller JR 3rd, Desprez PY, McAllister S, Soroceanu L, Debs RJ, Salomonis N, Schadendorf D, Cleaver JE, Kashani-Sabet M (2018) PHIP as a therapeutic target for driver-negative subtypes of melanoma, breast, and lung cancer. Proc Natl Acad Sci U S A 115(25):E5766–E5775
De Semir D, Nosrati M, Bezrookove V, Dar AA, Federman S, Bienvenu G, Venna S, Rangel J, Climent J, Meyer Tamguney TM, Thummala S, Tong S, Leong SP, Haqq C, Billings P, Miller JR 3rd, Sagebiel RW, Debs R, Kashani-Sabet M (2012) Pleckstrin homology domain-interacting protein (PHIP) as a marker and mediator of melanoma metastasis. Proc Natl Acad Sci U S A 109(18):7067–7072
Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, Sleckman BP, Nussenzweig A (2008) 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456(7221):529–533
Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C (2009) RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136(3):435–446
Farhang-Fallah J, Randhawa VK, Nimnual A, Klip A, Bar-Sagi D, Rozakis-Adcock M (2002) The pleckstrin homology (PH) domain-interacting protein couples the insulin receptor substrate 1 PH domain to insulin signaling pathways leading to mitogenesis and GLUT4 translocation. Mol Cell Biol 22(20):7325–7336
Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A (2004) H2AX: the histone guardian of the genome. DNA Repair 3(8–9):959–967
Fishman-Lobell J, Rudin N, Haber JE (1992) Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol 12(3):1292–1303
Huen MS, Sy SM, van Deursen JM, Chen J (2008) Direct interaction between SET8 and proliferating cell nuclear antigen couples H4–K20 methylation with DNA replication. J Biol Chem 283(17):11073–11077
Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD (2004) Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432(7015):406–411
Indig FE, Partridge JJ, von Kobbe C, Aladjem MI, Latterich M, Bohr VA (2004) Werner syndrome protein directly binds to the AAA ATPase p97/VCP in an ATP-dependent fashion. J Struct Biol 146(1–2):251–259
Indig FE, Rybanska I, Karmakar P, Devulapalli C, Fu H, Carrier F, Bohr VA (2012) Nucleolin inhibits G4 oligonucleotide unwinding by Werner helicase. PLoS ONE 7(6):e35229
Jang SM, Nathans JF, Fu H, Redon CE, Jenkins LM, Thakur BL, Pongor LS, Baris AM, Gross JM, O’Neill MJ, Indig FE, Cappell SD, Aladjem MI (2020a) The RepID-CRL4 ubiquitin ligase complex regulates metaphase to anaphase transition via BUB3 degradation. Nat Commun 11(1):24
Jang SM, Redon CE, Aladjem MI (2018a) Chromatin-bound Cullin-ring ligases: regulatory roles in DNA replication and potential targeting for cancer therapy. Front Mol Biosci 5:19
Jang SM, Redon CE, Thakur BL, Bahta MK, Aladjem MI (2020b) Regulation of cell cycle drivers by Cullin-RING ubiquitin ligases. Exp Mol Med 52(10):1637–1651
Jang SM, Zhang Y, Utani K, Fu H, Redon CE, Marks AB, Smith OK, Redmond CJ, Baris AM, Tulchinsky DA, Aladjem MI (2018b) The replication initiation determinant protein (RepID) modulates replication by recruiting CUL4 to chromatin. Nat Commun 9(1):2782
Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH (2011) Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 333(6051):1895–1898
Ji X, Dadon DB, Abraham BJ, Lee TI, Jaenisch R, Bradner JE, Young RA (2015) Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proc Natl Acad Sci U S A 112(12):3841–3846
Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS (2007) The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol 179(7):1337–1345
Kim JJ, Lee SY, Gong F, Battenhouse AM, Boutz DR, Bashyal A, Refvik ST, Chiang CM, Xhemalce B, Paull TT, Brodbelt JS, Marcotte EM, Miller KM (2019) Systematic bromodomain protein screens identify homologous recombination and R-loop suppression pathways involved in genome integrity. Genes Dev 33(23–24):1751–1774
Kim Y, Starostina NG, Kipreos ET (2008) The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev 22(18):2507–2519
Kuo CY, Li X, Stark JM, Shih HM, Ann DK (2016) RNF4 regulates DNA double-strand break repair in a cell cycle-dependent manner. Cell Cycle 15(6):787–798
Kurat CF, Yeeles JTP, Patel H, Early A, Diffley JFX (2017) Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol Cell 65(1):117–130
Li S, Francisco AB, Han C, Pattabiraman S, Foote MR, Giesy SL, Wang C, Schimenti JC, Boisclair YR, Long Q (2010) The full-length isoform of the mouse pleckstrin homology domain-interacting protein (PHIP) is required for postnatal growth. FEBS Lett 584(18):4121–4127
Liu L, Lee S, Zhang J, Peters SB, Hannah J, Zhang Y, Yin Y, Koff A, Ma L, Zhou P (2009) CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell 34(4):451–460
Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412(6846):557–561
Ma Y, Lu H, Schwarz K, Lieber MR (2005) Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle 4(9):1193–1200
Machida YJ, Hamlin JL, Dutta A (2005) Right place, right time, and only once: replication initiation in metazoans. Cell 123(1):13–24
Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, Sixma TK, Richard S (2012) RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J 31(8):1865–1878
Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB (2004) 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol 5(5):481–487
Meerang M, Ritz D, Paliwal S, Garajova Z, Bosshard M, Mailand N, Janscak P, Hubscher U, Meyer H, Ramadan K (2011) The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat Cell Biol 13(11):1376–1382
Mehta A, Haber JE (2014) Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol 6(9):a016428
Meyer H, Weihl CC (2014) The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J Cell Sci 127(Pt 18):3877–3883
Morgan MAJ, Rickels RA, Collings CK, He X, Cao K, Herz HM, Cozzolino KA, Abshiru NA, Marshall SA, Rendleman EJ, Sze CC, Piunti A, Kelleher NL, Savas JN, Shilatifard A (2017) A cryptic Tudor domain links BRWD2/PHIP to COMPASS-mediated histone H3K4 methylation. Genes Dev 31(19):2003–2014
Nalabothula N, Indig FE, Carrier F (2010) The nucleolus takes control of protein trafficking under cellular stress. Mol Cell Pharmacol 2(5):203–212
Partridge JJ, Lopreiato JO Jr, Latterich M, Indig FE (2003) DNA damage modulates nucleolar interaction of the Werner protein with the AAA ATPase p97/VCP. Mol Biol Cell 14(10):4221–4229
Pfeiffer P, Goedecke W, Obe G (2000) Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis 15(4):289–302
Podcheko A, Northcott P, Bikopoulos G, Lee A, Bommareddi SR, Kushner JA, Farhang-Fallah J, Rozakis-Adcock M (2007) Identification of a WD40 repeat-containing isoform of PHIP as a novel regulator of beta-cell growth and survival. Mol Cell Biol 27(18):6484–6496
Smith KD, Fu MA, Brown EJ (2009) Tim-Tipin dysfunction creates an indispensable reliance on the ATR-Chk1 pathway for continued DNA synthesis. J Cell Biol 187(1):15–23
Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D (2009) The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136(3):420–434
Symington LS, Gautier J (2011) Double-strand break end resection and repair pathway choice. Annu Rev Genet 45:247–271
Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E (2010) The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 12(11):1086–1093
Tokuda E, Fujita N, Oh-hara T, Sato S, Kurata A, Katayama R, Itoh T, Takenawa T, Miyazono K, Tsuruo T (2007) Casein kinase 2-interacting protein-1, a novel Akt pleckstrin homology domain-interacting protein, down-regulates PI3K/Akt signaling and suppresses tumor growth in vivo. Cancer Res 67(20):9666–9676
Torrecilla, I., J. Oehler and K. Ramadan (2017). "The role of ubiquitin-dependent segregase p97 (VCP or Cdc48) in chromatin dynamics after DNA double strand breaks." Philos Trans R Soc Lond B Biol Sci 372(1731).
Van C, Yan S, Michael WM, Waga S, Cimprich KA (2010) Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J Cell Biol 189(2):233–246
van den Boom J, Wolf M, Weimann L, Schulze N, Li F, Kaschani F, Riemer A, Zierhut C, Kaiser M, Iliakis G, Funabiki H, Meyer H (2016) VCP/p97 extracts sterically trapped Ku70/80 rings from DNA in double-strand break repair. Mol Cell 64(1):189–198
Vaz B, Halder S, Ramadan K (2013) Role of p97/VCP (Cdc48) in genome stability. Front Genet 4:60
Wall ME, Wani MC (1995) Camptothecin and taxol: discovery to clinic–thirteenth Bruce F. Cain Memorial Award Lecture. Cancer Res 55(4):753–760
Wyrobek AJ, Schmid TE, Marchetti F (2005) Relative susceptibilities of male germ cells to genetic defects induced by cancer chemotherapies. J Natl Cancer Inst Monogr. 34:31–35
Yang C, Maiguel DA, Carrier F (2002) Identification of nucleolin and nucleophosmin as genotoxic stress-responsive RNA-binding proteins. Nucleic Acids Res 30(10):2251–2260
Zhang H, Wang Q, Kajino K, Greene MI (2000) VCP, a weak ATPase involved in multiple cellular events, interacts physically with BRCA1 in the nucleus of living cells. DNA Cell Biol 19(5):253–263
Zhang Y, Huang L, Fu H, Smith OK, Lin CM, Utani K, Rao M, Reinhold WC, Redon CE, Ryan M, Kim R, You Y, Hanna H, Boisclair Y, Long Q, Aladjem MI (2016) A replicator-specific binding protein essential for site-specific initiation of DNA replication in mammalian cells. Nat Commun 7:11748
Acknowledgement
This study was supported by the Intramural Research Program of the NIH, Center for Cancer Research, National Cancer Institute, and by the Intramural Research Program of the NIH, National Institute on Aging.
Author information
Authors and Affiliations
Contributions
SMJ and MIA designed the study. SMJ, CER, HF and FEI designed and performed the experiments. SMJ and MIA wrote the manuscript. SMJ, CER, HF, FEI, and MIA revised the manuscript.
Corresponding authors
Ethics declarations
Human and animal rights
The study does not involve any with humans or animal experiments, and this study was performed following institutional and national guidelines.
Conflict of interests
Sang-Min Jang, Christophe E. Redon, Haiqing Fu, Fred E. Indig & Mirit I. Aladjem declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jang, SM., Redon, C.E., Fu, H. et al. RepID-deficient cancer cells are sensitized to a drug targeting p97/VCP segregase. Mol. Cell. Toxicol. 17, 141–149 (2021). https://doi.org/10.1007/s13273-021-00121-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13273-021-00121-0