Abstract
Background
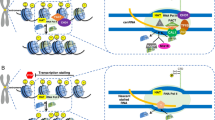
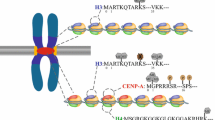
Centromeres are specialized chromosomal domains involved in kinetochore formation and faithful chromosome segregation. Despite a high level of functional conservation, centromeres are not identified by DNA sequences, but by epigenetic means. Universally, centromeres are typically formed on highly repetitive DNA, which were previously considered to be silent. However, recent studies have shown that transcription occurs in this region, known as centromeric-derived RNAs (cenRNAs). CenRNAs that contribute to fundamental aspects of centromere function have been recently investigated in detail. However, the distribution, behavior and contributions of centromeric transcripts are still poorly understood.
Objective
The aim of this article is to provide an overview of the roles of cenRNAs in centromere formation and function.
Methods
We describe the structure and DNA sequence of centromere from yeast to human. In addition, we briefly introduce the roles of cenRNAs in centromere formation and function, kinetochore structure, accurate chromosome segregation, and pericentromeric heterochromatin assembly. Centromeric circular RNAs (circRNAs) and R-loops are rising stars in centromere function. CircRNAs have been successfully identified in various species with the assistance of high-throughput sequencing and novel computational approaches for non-polyadenylated RNA transcripts. Centromeric R-loops can be identified by the single-strand DNA ligation-based library preparation technique. But the molecular features and function of these centromeric R-loops and circRNAs are still being investigated.
Conclusion
In this review, we summarize recent findings on the epigenetic regulation of cenRNAs across species, which would provide useful information about cenRNAs and interesting hints for further studies.




Similar content being viewed by others
References
Arabidopsis Genome I (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Arunkumar G, Melters DP (2020) Centromeric transcription: a conserved swiss-army knife. Genes 11:911
Bergmann JH, Rodríguez MG, Martins NMC, Kimura H, Kelly DA, Masumoto H et al (2011) Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 30:328–340
Blower MD (2016) Centromeric transcription regulates aurora-B localization and activation. Cell Rep 15:1624–1633
Boeger H, Griesenbeck J, Strattan JS, Kornberg RD (2003) Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11:1587–1598
Bouzinba-Segard H, Guais A, Francastel C (2006) Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci U S A 103:8709–8714
Braselton JB (1975) Ribonucleoprotein staining of Allium cepa kinetochores. Cytobiologie 12:148–151
Bury L, Moodie B, Ly J, Mckay LS, Miga KH, Cheeseman IM (2020) Alpha-satellite RNA transcripts are repressed by centromere-nucleolus associations. Elife 9:e59770
Camacho OV, Galan C, Rosowska KS, Ching R, Gamalinda M, Karabiber F, Velazquez IDLR, Engist B, Koschorz B, Shukeir N et al (2017) Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. Elife 6:e25293
Chan FL, Wong LH (2012) Transcription in the maintenance of centromere chromatin identity. Nucleic Acids Res 40:11178–11188
Chen ES, Saitoh S, Yanagida M, Takahashi K (2003) A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol Cell 11:175–187
Chen ES, Zhang K, Nicolas E, Hugh PC, Zofall M, Grewal SS (2008) Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451:734–737
Chen L, Zhang P, Fan Y et al (2018) Circular RNAs mediated by transposons are associated with transcriptomic and phenotypic variation in maize. New Phytol 217:1292–1306
Choi ES, Stralfors A, Castillo AG, Durand-Dubief M, Ekwall K, Allshire RC (2011) Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J Biol Chem 286:23600–23607
Choi ES, Strålfors A, Catania S, Castillo AG, Svensson JP, Pidoux AL, Ekwall K, Allshire RC (2012) Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-A (Cnp1) in fission yeast. PLoS Genet 8:e1002985
Choo KHA (2001) Domain organization at the centromere and neocentromere. Dev. Cell 1:165–177
Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH (2009) LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PloS Genet 5:e1000354
Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407–421
Collins KA, Castillo AR, Tatsutani SY, Biggins S (2005) De novo kinetochore assembly requires the centromeric histone H3 variant. Mol Biol Cell 16:5649–5660
Corless S, Höcker S, Erhardt S (2020) Centromeric RNA and its function at and beyond centromeric chromatin. J Mol Biol 432:4257–4269
Darbani B, Noeparvar S, Borg S (2016) Identification of circular RNAs from the parental genes involved in multiple aspects of cellular metabolism in Barley. Front Plant Sci 7:776
Dhatchinamoorthy K, Mattingly M, Gerton JL (2018) Regulation of kinetochore configuration during mitosis. Curr Genet 64:1197–1203
Du Y, Topp CN, Dawe RK (2010) DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet 6:e1000835
Dunleavy EM, Beier NL, Gorgescu W, Tang J, Costes SV, Karpen GH (2012) The cell cycle timing of centromeric chromatin assembly in Drosophila meiosis is distinct from mitosis yet requires CAL1 and CENP-C. PLoS Biol 10:e1001460
Fan X, Zhang X, Wu X, Guo H, Hu Y, Tang F, Huang Y (2015) Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol 16:148
Fang Y, Chen LF, Lin K, Feng YL, Zhang PY, Pan XC et al (2019) Characterization of functional relationships of R-loops with gene transcription and epigenetic modifications in rice. Genome Res 29:1287–1297
Feretzaki M, Pospisilova M, Fernandes RV, Lunardi T, Krejci L, Lingner J (2020) RAD51-dependent recruitment of TERRA ncRNA to telomeres through R-loops. Nature 587:303–308
Ferri F, Bouzinba-Segard H, Velasco G et al (2009) Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res 37:5071–5080
Frederic C, Craig JB (2020) Emerging roles for R-loop structures in the management of topological stress. J Biol Chem 3:4684–4695
Georg OM, Bobkov NG, Patrick H (2018) Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. J Cell Biol 217:1957–1972
Graf M et al (2017) Telomere length determines TERRA and R-loop regulation through the cycle. Cell 170:2–85
Grenfell AW, Heald R, Strzelecka M (2016) Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. J Cell Biol 214:133–141
Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA (2017) Transcription-replication conflict orientation modulates r-loop levels and activates distinct DNA damage responses. Cell 70:774–786
Hao YJ, Wang DP, Wu SH, Li X, Shao CW, Zhang P, Chen JY, Lim DH, Fu XD et al (2020) Active retrotransposons help maintain pericentromeric heterochromatin required for faithful cell division. Genome Res 30:1570–1582
Heieh CL, Xia J, Lin HF (2020) MIWI prevents aneuploidy during meiosis by cleaving excess satellite RNA. EMBO J 39:e103614
Henikoff S, Talbert PB (2020) What makes a centromere? Exp. Cell Res. 389: 111895 Henikoff S, Ahmad K, Malik HS (2001) The Centromere Paradox: Stable Inheritance with Rapidly Evolving DNA. Science 293:1098–1110
Henikoff S, Buell CR, Jiang J (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36:138–145
Hill A, Bloom K (1987) Genetic manipulation of centromere function. Mol Cell Biol 7:2397–2405
Ishikura S, Nakabayashi K, Nagai M, Tsunoda T, Shirasawa S (2020) ZFAT binds to centromeres to control noncoding RNA transcription through the KAT2B-H4K8ac-BRD4 axis. Nuclei Acids Res 48:10848–10866
Ivanov S, Memczak E, Wyler F, Torti HT, Porath MR, Orejuela M, Piechotta EY, Levanon M, Landthaler C, Dieterich N, Rajewsky, (2015) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10:170–177
Jansen LE, Black BE, Foltz DR, Cleveland DW (2007) Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176:795–805
Jiang J, Birchler JA, Parrott WA, Dawe RK (2003) A molecular view of plant centromeres. Trends Plant Sci 8:570–575
Kabeche L, Nguyen HD, Buisson R, Zou L (2018) A Mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science 359:108–114
Kobayashi N, Suzuki Y, Schoenfeld LW, Müller CA et al (2015) Discovery of an unconventional centromere in budding yeast redefines evolution of point centromeres. Curr Biol 3:2026–2033
Lee HR, Neumann P, Macas J, Jiang J (2006) Transcription and evolutionary dynamics of the centromeric satellite repeat CentO in rice. Mol Biol Evol 23:2505–2520
Lefrançois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, Gerstein M, Snyder M (2009) Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics 10:37
Lermontova I, Fuchs J, Schubert V, Schubert I (2007) Loading time of the centromeric histone H3 variant differs between plants and animals. Chromosoma 116:507–510
Lermontova I, Rutten T, Schubert I (2011) Deposition, turnover, and release of CENH3 at Arabidopsis centromeres. Chromosoma 120:633–640
Ling YH, Wing K, Yuen Y (2019) Centromeric non-coding RNA as a hidden epigenetic factor of the point centromere. Curr Genet 65:1165–1171
Lippman Z, Martienssen R (2004) The role of RNA interference in heterochromatic silencing. Nature 431:364–370
Liu Y, Su H, Zhang J, Liu Y, Feng C, Han FP (2020) Back-spliced RNA from retrotransposon binds to centromere and regulates centromeric chromatin loops in maize. PLoS Biol 18:e3000582
Lu WT, Hawley BR, Skalka GL, Baldock RA, Smith EM, Bader AS, Malewicz M, Watts FZ, Wilczynska A, Bushell M (2018) Drosha drives the formation of DNA: RNA hybrids around DNA break sites to facilitate DNA repair. Nat Commun 9:532
Lv J, Yu K, Wei J, Gui H, Liu CX, Liang D, Wang YL et al (2020) Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3. Nat Biotechnol. 38:1397–1401
Maison C, Quivy JP, Probst AV, Almouzni G (2010) Heterochromatin at mouse pericentromeres: a model for de novo heterochromatin formation and duplication during replication. Cold Spring Harb Symp Quant Biol 75:155–165
Maldonado R, Schwartz U, Silberhorn E, Längst G (2019) Nucleosomes Stabilize ssRNA-dsDNA Triple Helices in Human Cells. Mol Cell 73:1243–1254
May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA (2005) Differential regulation of strand-specific transcripts from arabidopsis centromeric satellite repeats. PLoS Genet 1:e79
Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH (2011) Assembly of drosophila centromeric chromatin proteins during mitosis. PLoS Genet 7:e1002068
Memczak S, Jens S, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338
Miga KH, Shoshani O, Asron A, McMahon MA, Lee AY et al (2019) DNA replication acts as an error correction mechanism to maintain centromere identity by restricting CENP-A to centromeres. Nat Cell Biol 21:743–754
Mishra PK, Chakrabortyb K, Yehc E, Feng WY, Bloomc KS, Basraia M (2020) R-loops at centromeric chromatin contribute to defects in kinetochore integrity and chromosomal instability in budding yeast. Mol Biol Cell 1:mbcE20060379
MuNulty SM et al (2017) Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Dev Cell 42:226–240
Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Nechemia-Arbely Y, Ideue T, Cho Y, Nishimura K, Tani T (2014) Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes Cells 19:528–538
Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-lewis S, Larionov V, Earnshaw WC, Masumoto H (2008) Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell 14:507–522
Neumann P, Yan H, Jiang J (2007) The centromeric retrotransposons of rice are transcribed and differentially processed by RNA interference. Genetics 176:749761
Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SIS (2007) Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol 14:372–380
Ohkuni K, Kitagawa K (2011) Endogenous Transcription at the Centromere Facilitates Centromere Activity in Budding Yeast. Curr. Biol. 21:1695–1703
Ólafsson G, Thorpe PH (2020) Polo kinase recruitment via the constitutive centromere-associated network at the kinetochore elevates centromeric RNA. PLoS Genet 18:e1008990
Path K, Mlynarcayk-Evans S, Nusinow D, Panning B (2002) Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet 36:233–278
Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC (1995) The centromere: hub of chromosomal activities. Science 270:1591–1594
Quénet D, Dalal Y (2014) A long non-coding RNA is required for targeting centromeric protein a to the human centromere. Elife 3:e03254
Reinhart BJ, Bartel DP (2002) Small RNAs correspond to centromere heterochromatic repeats. Science 297:1831
Rieder CL (1979) Ribonucleoprotein staining of centrioles and kinetochores in newt lung cell spindles. J Cell Biol 80:1–9
Rošić S, Köhler F, Erhardt S (2014) Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J Cell Biol 207:335–349
Saffery R, Sumer H, Hassan S, Wong LH, Craig JM, Todokoro K, Anderson M, Safford A, Choo KHA (2003) Transcription within a functional human centromere. Mol Cell 12:509–516
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7:e30733
Schoeftner S, Blasco MA (2009) A “higher order” of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J 28:2323–2336
Schubert V, Lermontova I, Shubert I (2014) Loading of the centromeric histone H3 variant during meiosis – how does it differ from mitosis? Chromosoma 123: 491–7 Shao JJ, Wang LQ, Liu XY, Yang M, Chen HM, Wu B, Liu C (2019) Identification and characterization of circular RNAs in Ganoderma lucidum. Sci Rep 9:16522
Shuhei I, Kazuhiko N, Masayoshi N, Toshiyuki T, Senji S (2020) ZFAT binds to centromeres to control noncoding RNA transcription through the KAT2B–H4K8ac–BRD4 axis. Nucleic Acids Res 4:10848–10966
Stimpson KM, Sullivan BA (2010) Epigenomics of centromere assembly and function. Curr Opin Cell Biol 22:772–780
Su H, Liu YL et al (2016) Dynamic chromatin changes associated with de novo centromere formation in maize euchromatin. Plant J 88:854–866
Su H, Liu YL et al (2019) Centromere Satellite Repeats Have Undergone Rapid Changes in Polyploid Wheat Subgenomes. Plant Cell 31:2015–2051
Sullivan KF (2001) A solid foundation: functional specialization of centromeric chromatin. Curr Opin Genet 11:182–188
Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11:1076–1083
Tallbert PB, Henikoff S (2018) Transcribing centromeres: noncoding RNAs and kinetochore assembly. Trends Genet 34:587–599
Topp CN, Zhong CX, Dawe RK (2004) Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci USA 101:15986–15991
Unoki M, Sharif J, Saito YC et al (2020) CDCA7 and HELLS suppress DNA:RNA hybrid-associated DNA damage at pericentromeric repeats. Sci Rep 10:17865
Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI et al (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672–676
Volpe TA, Kider C, Hall IM, Teng G, Grewal SIS, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 Lysine-9 Methylation by RNAi. Sci 297:1833–1837
Wang Z, Liu Y, Li D et al (2017a) Identification of circular RNAs in Kiwifruit and their species-specific response to bacterial canker pathogen invasion. Front Plant Sci 8:413
Wang Y, Yang M, Wei S et al (2017b) Identification of circular RNAs and their targets in leaves of Triticum aestivum L under dehydration stress. Front Plant Sci 7:224
Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC (2014) Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neuralaccumulation. Cell Rep. 9:1966–1980
Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, Choo KH (2007) Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res 17:1146–1160
Xu W, Xu H, Li K, Fan YX, Liu Y, Yang XR, Sun QW (2017) The R-loop is a common chromatin feature of the Arabidopsis genome. Nat Plants 3:704–714
Yamagishi Y, Sakuno T, Goto Y, Watanabe Y (2014) Kinetochore composition and its function: lessons from yeasts. FEMS Microbiol Rev 38:185–200
Ye CY, Liu C, Liu C, Zhu QH, Fan LJ (2015) Widespread noncoding circular RNAs in plants. New Phytol 208:88–95
Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR (2003) R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 4:442–451
Zeng RF, Zhou JJ, Hu CG et al (2018) Transcriptome-wide identification and functional prediction of novel and flowering-related circular RNAs from trifoliate orange (Poncirus trifoliata L. Raf.). Planta 247:1191–1202
Zhang X, Ma X, Ning L, Li Z, Zhao K, Li K, He J, Yin D (2019) Genome-wide identification of circular RNAs in peanut (Arachis hypogaea L). BMC Genomics 20:653
Zhao T, Wang L, Li S et al (2017a) Characterization of conserved circular RNA in polyploid Gossypium species and their ancestors. FEBS Lett 591:3660–3669
Zhao W, Cheng Y, Zhang C et al (2017b) Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci Rep 7:5636
Zuo J, Wang Q, Zhu B et al (2016) Deciphering the roles of circRNAs on chilling injury in tomato. Biochem Biophys Res Commun 479:132–138
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31920103006 and 31630049) and a National Science Foundation plant genome grant (IOS-1444514).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Q., Liu, Y., Shi, Q. et al. Emerging roles of centromeric RNAs in centromere formation and function. Genes Genom 43, 217–226 (2021). https://doi.org/10.1007/s13258-021-01041-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-021-01041-y