Abstract
Background
Respiratory diseases in pigs are the main health concerns for swine producers. Similar to the diseases in human and other animals, respiratory diseases are primary related to morbidity and are the result of infection with bacteria, viruses, or both. B. bronchiseptica causes serious respiratory diseases in the swine airway track. However, the B. bronchiseptica-specific bacteriophage has diverse advantages such as decreasing antibiotic overuse and possible therapeutic potential against bacteria.
Objective
The objects of this study were to investigate the therapeutic effect of specific B. bronchiseptica bacteriophages and to identify genes related to bacteriophage signaling utilizing RNA microarrays in swine nasal turbinate cells.
Methods
Bor-BRP-1 phages were applied 24 h prior to B.bronchiseptica infection (1 × 107 cfu/ml) at several concentrations of bacterial infection. Cells were incubated to detect cytokines and 24 h to detect mucin production. And real-time quantitative PCR was performed to examine related genes expression. To determine the change of total gene expression based on B.bronchiseptica and Bor-BRP-1 treatment, we performed RNA sequencing experiments.
Results
The results showed that B. bronchiseptica induced increased expression of several inflammatory genes such as IL-1β, IL-6, and Muc1 in a dose-dependent manner. However, Bor-BRP-1 induced reduction of gene expression compared to the B. bronchiseptica induction group. In addition, microarrays detected Bor-BRP-1-altered inflammatory gene expression against B. bronchiseptica, reducing B. bronchiseptica-induced airway inflammation in swine epithelial cells.
Conclusion
These results suggest that the specific bacteriophage has a therapeutic potential to defend against B. bronchiseptica infection by altering inflammatory gene expression profiles.
Similar content being viewed by others
Introduction
Bordetella bronchiseptica is a widespread gram-negative coccobacillus that causes respiratory diseases in wide range of mammalian species. Currently, it is a well-recognized etiological agent in many domestic and wild animal diseases including tracheobronchitis, conjunctivitis, rhinitis, and pneumonia (Garbal et al. 2016; Petrovic et al. 2017). In particular, it is one of the primary causative agents of atrophic rhinitis and bronchopneumonia in swine (Moore and Kern 2001; Prüller et al. 2015; Chen et al. 2019). The characteristic lesion of atrophic rhinitis is atrophy of the nasal septum and turbinate, leading to facial deformation (Petrovic et al. 2017; De Jong and Nielsen 1990; Brockmeier et al. 2002). This impairs swine feeding and causes a reduction in growth performance resulting in great economic losses in the pig-rearing industry.
B. bronchiseptica is often isolated in mixtures with other bacterial or viral pathogens such as porcine reproductive and respiratory syndrome (PRRS) virus, porcine respiratory coronavirus, swine influenza virus (SIV), Haemophilus parasuis, Pasteurella multocida, and Streptococcus suis, from respiratory diseases in swine (Chen et al. 2019; Park et al. 2018; King et al. 2002). B. bronchiseptica infection predisposes the upper respiratory tract to colonization with secondary pathogens including P. multocida and S. suis (Brockmeier 2004; Pósa et al. 2011). P. multocida inefficiently colonizes by itself, while B. bronchiseptica acts as a precursor for infection with P. multocida. In addition, pre-inoculation with B. bronchiseptica prior to S. suis resulted in elevated isolation of S. suis as well as increased incidence of pneumonia and mortality in pigs (Mengelers et al. 1989; Vecht et al. 1992). Presentation of B. bronchiseptica-infected mammalian animals can vary from asymptomatic cases to severe systemic infections. Generally, this infection leads to life-long chronic colonization in the host. Once infected, the infection is rarely eliminated from the host (Irie et al. 2005).
Bacteriophages are tiny microorganisms that infect bacteria and proliferate within bacterial cells. After proliferation, the phages escape from the bacteria by destroying the bacterial cell wall (Forde and Fitzgerald 1999; Hermoso et al. 2007; Pepin et al. 2008). Phages are very specific and can only selectively kill the target bacteria, not affecting other bacteria such as normal flora in animals (Leverentz et al. 2003; Rees and Dodd 2006). Although B. bronchiseptica was first reported in 1911, several studies have been performed with its bacteriophages. Several reports have revealed that B. bronchiseptica is isolated through antibiotic resistance to β-lactams, macrolides, etc. (Petrovic et al. 2017; Prüller et al. 2015; Biswas et al. 2002; Dayao et al. 2014; Niemann et al. 2018). Previously, we reported that Bor-BRP-1 down-regulated B. bronchiseptica-induced inflammation (Park et al. 2018). However, the physiological mechanism by which Bor-BRP-1 suppresses B. bronchiseptica-induced inflammation remains unidentified.
In this report, we demonstrated that B. bronchiseptica-induced inflammatory RNA expression can be effectively controlled by treatment with B. bronchiseptica-specific bacteriophage Bor-BRP-1 in vitro. Therefore, bacteriophages may function to suppress bacterial inflammation and may be a potential therapeutic candidate to eliminate B. bronchiseptica.
Materials and methods
Materials
Swine nasal turbinate (PT-K75) cells were purchased from ATCC (CRL-2528; Manassas, VA) and cultured as previously described (Park et al. 2018). Cells were prepared at 5 × 105 cells/well in 6-well plates and incubated for approximately one day to reach full confluence. Cells were serum starved for 24 h before treatment.
bronchiseptica culture and Bor-BRP-1 bacteriophage production
B. bronchiseptica strains (BB-01) were grown on Brain Heart Infusion (BHI) broth in a shaking incubator at 37 °C without antibiotics. The bacterial concentration was adjusted to 5 × 109 cfu/ml based on OD 600 nm. According to a previous study, the inoculum of B. bronchiseptica to PT-K75 cells was selected at 1 × 107 cfu/ml. Bor-BRP-1 bacteriophage was produced based on a previous study (Park et al. 2018). Bor-BRP-1 phages were applied 24 h prior to B. bronchiseptica-infection (1 × 107 cfu/ml) at 10 × concentration. One day after bacterial infection, the cells were lysed with TRIzol for RNA extraction.
Real-time quantitative PCR
Real-time PCR was carried out on an ABI 7300 Real-Time PCR system (Applied Biosystems, Foster City, CA) with TB Green™ Premix Ex Taq™ II (RR820A, Takara, Japan). Reactions were performed in a total volume of 20 µl including 10 µl of 2 × SYBR Green PCR Master Mix, 300 nM of each primer, and 1 µl of the previously reverse-transcribed cDNA template. The following primers were used: porcine IL-1β, forward 5′-GAGCATCAGGCAGATGGTGT -3′ and reverse 5′- CAAGGATGATGGGCTCTTCTTC -3′; porcine IL-6, forward 5′- GCTGCTTCTGGTGATGGCTACTGCC -3′ and reverse 5′- TGAAACTCCACAAGACCGGTGGTGA -3′, porcine Muc1 forward 5′- GTGCCGACGAAAGAACTG -3′ and reverse 5′- TGCCAGGTTCGAGTAAGAG -3′, porcine GAPDH, forward 5′- AAGGAGTAAGAGCCCCTGGA -3′ and reverse 5′- TCTGGGATGGAAACTGGAAG -3′. All reactions were performed in triplicate. The relative quantity of mRNA was obtained using a comparative cycle threshold method and was normalized using GAPDH as an endogenous control.
Quant-Seq microarray
Total RNA was quantified with an Agilent 2100 bioanalyzer using the RNA 6000 Nano Chip (Agilent Technologies, Amstelveen, The Netherlands). RNA quantification was determined using an ND-2000 Spectrophotometer (Thermo Inc., DE, USA). The library was constructed using a QuantSeq 3′ mRNA-Seq Library Prep Kit (Lexogen, Inc., Austria) according to the manufacturer’s instructions. In brief, 500 ng of total RNA was prepared, an oligo-dT primer containing an Illumina-compatible sequence at its 5′ end was hybridized to the RNA, and reverse transcription was applied. After RNA template degradation, second-strand synthesis was initiated by a random primer containing an Illumina-compatible linker sequence at its 5′ end. The double-stranded library was purified using magnetic beads for removal of all reaction components. The library was amplified to add the complete adapter sequences required for cluster generation. The finished library was purified from PCR components. High-throughput sequencing was performed as single-end 75 sequencing using NextSeq 500 (Illumina, Inc., USA). For data analysis, QuantSeq 3′ mRNA-Seq reads were aligned using Bowtie2 (Langmead and Salzberg 2012). Bowtie2 indices were either generated from the genome assembly sequence or the representative transcript sequences for aligning to the genome and transcriptome. The alignment file was used for assembling transcripts, estimating their abundances, and detecting differential expression of genes. Differentially expressed genes were determined based on counts from unique and multiple alignments using coverage in Bedtools (Quinlan 2010). The Read Count data were processed based on the quantile normalization method using EdgeR within R using Bioconductor (Gentleman et al. 2004). Gene classification was based on searches in Database for Annotation, Visualization, and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/) and Medline databases (http://www.ncbi.nlm.nih.gov/). A Porcine Quant-Seq microarray was performed using the customized service provided by eBiogen Inc, (Seoul, South Korea).
Statistical analysis
Data are presented as the mean ± SD of at least three independent experiments. Where appropriate, statistical differences among the groups were assessed by the ANOVA test. Differences with p value less than 0.05 was considered statistically significant.
Results
Bor-BRP‑1 significantly inhibited B. bronchiseptica‑induced airway inflammation in PT‑K75 cells
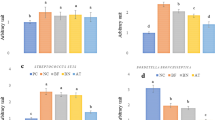
First, we performed cell viability assays and confirmed that neither B. bronchiseptica nor Bor-BRP-1 exhibited cytotoxicity in a dose- or time-dependent manner (data not shown). B. bronchiseptica administration induced inflammatory cytokine secretion and mucin gene expression to control the respiratory microenvironment in our previous study (Park et al. 2018). In addition, Bor-BRP‑1 was challenged for 24 h in prior B. bronchiseptica infections. Bor-BRP-1 significantly decreased B. bronchiseptica-induced IL-1β, IL-6, and Muc1 expression (Fig. 1a, b). Muc1 is known as an anti-inflammatory mucin (Kato et al. 2017). Since the airway inflammatory environment was decreased by Bor-BRP-1, Muc1 gene expression decreased proportional to the status of anti-inflammation. The results indicate that Bor-BRP‑1 dramatically suppressed the B. bronchiseptica-induced physiological phenomena by eliminating the bacteria.
Bor-BRP-1 bacteriophage significantly decreased IL-1β, IL-6, and Muc1 gene expression in PT-K75 cells. The indicated cfu/ml of B. bronchiseptica and Bor-BRP-1 phages was administrated to confluent PT-K75 cells in 6-well plates for either 4 h (for cytokines) or 24 h (for mucin). RNA and cell lysates were isolated for specific quantitative real time-PCR (a) and ELISA (b). The figures are representative of three independent experiments. *p < 0.05 compared with B. bronchiseptica only
Gene expression in response to Bor-BRP-1 treatment and B. bronchiseptica infection
To examine the impact of gene expression according to Bor-BRP-1 treatment and B. bronchiseptic infection, RNA sequencing was performed on each group of PT-K 75 cells. To investigate the effect of Bor-BRP-1 on B. bronchiseptica-infected cells, we classified the four sample groups as follows: (1) Control group with no treatment, (2) B. bronchiseptica infection group, (3) Bor-BRP-1 treatment group, and (4) Bor-BRP-1 treatment prior to B. bronchiseptica infection group. Cells were lysed using TRIzol to analyze the altered genes through a microarray.
Following normalization, a differential comparison of the expression profiles was performed. The normalized read counts were greater than 4, and the fold change (FC) was greater than 2, with a p value < 0.05. A total of 304 DEGs was identified. Firstly, hierarchical clustering of RNA-sequencing data was conducted to view the global features of the data. The samples from each treatment group were normalized with a z score, and the statistical significance for differential expression was displayed among the groups (Fig. 2). Upregulation is indicated in red, and downregulation of a gene is indicated in blue. The results suggest that each comparison involved dramatic up- or downregulation according to treatment. These results showed a dramatic increase in the group of B. bronchiseptica infection but also a significant decrease in the group of Bor-BRP-1 treatment prior to B. bronchiseptica infection.
Hierarchical clustering and heat map of RNA-seq data. Clustering of RNA-seq data generated in response to Control, B. bronchiseptica infection, Bor-BRP-1 treatment without B. bronchiseptica infection, and Bor-BRP-1 treatment 24 h prior to B. bronchiseptica induction groups. The heat map was generated using genes differentially expressed by at least twofold under two or more conditions. Downregulation of a gene is indicated in blue, and upregulation is indicated in red. Colors changing from blue to red indicate the differences in gene expression among the groups of control. Each row represents a single gene, and each column represents a group (color figure online)
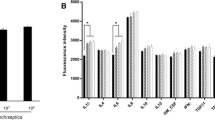
Among a number of genes, we selected several inflammation-related genes that exhibited representative changes depending on the analysis (Fig. 3). The results showed remarkable upregulation in the group of B. bronchiseptica infection but significant downregulation in the group with Bor-BRP-1 bacteriophage treatment prior to bacterial infection. These results agree with those in Fig. 1. Taken together, these findings indicate that Bor-BRP-1 suppressed B. bronchiseptica-induced airway inflammation by altering the inflammation-related mRNA profile in swine epithelial cells.
Selected genes from RNA sequencing. The genes resulted in marked upregulation in the group of B. bronchiseptica induction, whereas dramatic downregulation was exhibited in Bor-BRP-1 bacteriophage treatment prior to bacterial infection. ACSM4, acyl-CoA synthetase medium-chain family member 4; RAB32, RAB32, member RAS oncogene family; GPN3, GPN-loop GTPase 3; BMPR1A, bone morphogenetic protein receptor type 1A; SERINC2, serine incorporator 2; LRP8, LDL receptor related protein 8; MRPS18B, mitochondrial ribosomal protein S18B; MPZL2, myelin protein zero like 2; MOSPD2, motile sperm domain containing 2. *p < 0.05 compared with the control; **p < 0.05 compared with B. bronchiseptica only
Functional enrichment analysis of DEGs
To identify the changes in biological features due to bacterial infection and phage treatment, the identified DEGs were assembled and mapped into DAVID for functional enrichment analysis (Table 1). The DEGs were enriched mainly in biological processes associated with immune response. For further details regarding the identified DEGs, pathways were analyzed using KEGG. Pathway analysis showed that the metabolic pathways of PI3K-Akt signaling and TNF signaling were significantly enriched among the DEGs (p < 0.05, Table 2). This indicates that signaling of Bor-BRP-1 mainly affected the PI3K-Akt pathway, TNF signaling, and AMPK signaling to suppress specific B. bronchiseptica-induced inflammatory signaling in swine epithelial cells. Therefore, Bor-BRP-1 may be a potential therapeutic candidate to eliminate B. bronchiseptica.
Discussion
B. bronchiseptica-induced respiratory disease creates several serious problems for swine producers (Prüller et al. 2015; Nicholson et al. 2012, 2014). In this study, we evaluated the therapeutic potential of the specific bacteriophage Bor-BRP-1 on B. bronchiseptica-infected swine nasal turbinate cells. To the best of our knowledge, this is the first study that suggests alternation of micro RNA expression profiles in the Bor-BRP-1 bacteriophage effect on B. bronchiseptica-infected cells. Treatment of Bor-BRP-1 for 24 h prior to bacterial infection showed significant inhibition of IL-1β and IL-6 secretion as well as Muc1 gene expression. Furthermore, we confirmed bacteriophage suppression of several genes of bacterial-induced increased expression by microarray analysis. The results of the study indicate that Bor-BRP-1 has therapeutic potential against B. bronchiseptica-associated respiratory disease.
As seen in Fig. 1, IL-1β, IL-6, and Muc1 gene expression was upregulated in the B. bronchiseptica induction group, whereas the genes were decreased in a dose-dependent manner when the Bor-BRP-1 phage was applied for 24 h prior to B. bronchiseptica infection. This is interesting because IL-1β and IL-6 are important mediators of inflammation even though Muc1 is a membrane-bound mucin that serves an anti-inflammatory role (Kato et al. 2017). It is implied that Bor-BRP-1 could suppress release of proinflammatory cytokines as well as anti-inflammatory responses. We still do not fully understand the mechanism by which the specific phage controls bacteria-induced airway inflammation in swine airway epithelial cells. Therefore, this study mainly focused on identifying the action mechanism of Bor-BRP-1.
We characterized micro RNA expression profiles known as epigenetic components that modulate gene expression post-transcriptionally. The hierarchical clustering map with 272 genes indicated differentially expressed gene cross-groups from downregulation to upregulation (Fig. 2). Microarray revealed and qPCR confirmed increased gene expression in the B. bronchiseptica-infected group as well as decreased gene expression in the Bor-BRP-1-pretreated group, as shown in Figs. 1 and 3. The result showed that the genes related to inflammation exhibited altered expression in the control, B. bronchiseptica induction, and Bor-BRP-1 treatment prior to B. bronchiseptica induction groups. In addition, the selected genes exhibited similar traits assumed to be affected by the specific bacteriophage to suppress B. bronchiseptica-induced intracellular changes by killing the bacteria.
There are certain limitations in our study. First, we assessed the cellular viability of the host cells but could not confirm biosafety of the bacteriophage for clinical use. Second, we verified a protective effect of the bacteriophage as pretreatment to bacterial infection, but we did not evaluate the bacteriophage effect after bacterial infection. Further studies need to be performed to determine whether the bacteriophage could eliminate the bacteria after infection with or without antibiotic withdrawal.
In summary, we identified a protective effect of Bor-BRP-1 in B. bronchiseptica-infected swine nasal turbinate cells when applied 24 h prior to bacterial infection. Importantly, Bor-BRP-1 did not affect the viability of the host cells and could be a powerful alternative medicine to B. bronchiseptica-induced airway inflammation. Future studies will be performed to identify the intracellular mechanism of bacterial elimination according to phage challenge.
References
Biswas B, Adhya S, Washart P, Paul B, Trostel AN, Powell B, Carlton R, Merril CR (2002) Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect Immun 70:204–210
Brockmeier SL (2004) Prior infection with Bordetella bronchiseptica increases nasal colonization by Haemophilus parasuis in swine. Vet Microbiol 99:75–78
Brockmeier SL, Register KB, Magyar T, Lax AJ, Pullinger GD, Kunkle RA (2002) Role of the dermonecrotic toxin of Bordetella bronchiseptica in the pathogenesis of respiratory disease in swine. Infect Immun 70:481–490
Chen Y, Yang L, Sun E, Song J, Wu B (2019) Characterisation of a newly detected bacteriophage infecting Bordetella bronchiseptica in swine. Arch Virol 164:33–40
Dayao DA, Gibson JS, Blackall PJ, Turni C (2014) Antimicrobial resistance in bacteria associated with porcine respiratory disease in Australia. Vet Microbiol 171:232–235
De Jong MF, Nielsen JP (1990) Definition of progressive atrophic rhinitis. Vet Rec 126:93
Forde A, Fitzgerald GF (1999) Bacteriophage defence systems in lactic acid bacteria. Antonie Van Leeuwenhoek 76:89–113
Garbal M, Adaszek Ł, Łyp P, Frymus J, Winiarczyk M, Winiarczyk S (2016) Occurence of Bordetella bronchiseptica in domestic cats with upper respiratory tract infections. Pol J Vet Sci 19:353–358
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80
Hermoso JA, Garcia JL, Garcia P (2007) Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr Opin Microbiol 10:461–472
Irie Y, O’toole GA, Yuk MH (2005) Pseudomonas aeruginosa rhamnolipids disperse Bordetella bronchiseptica biofilms. FEMS Microbiol Lett 250:237–243
Kato K, Lillehoj EP, Lu W, Kim KC (2017) MUC1: the first respiratory mucin with an anti-inflammatory function. J Clin Med 6:E110
King SJ, Leigh JA, Heath PJ, Luque I, Tarradas C, Dowson CG, Whatmore AM (2002) Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J Clin Microbiol 40:3671–3680
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Leverentz B, Conway WS, Camp MJ, Janisiewicz WJ, Abuladze T, Yang M, Saftner R, Sulakvelidze A (2003) Biocontrol of Listeria monocytogenes on fresh-cut produce by treatment with lytic bacteriophages and a bacteriocin. Appl Environ Microbiol 69:4519–4526
Mengelers MJ, van Klingeren B, van Miert AS (1989) In vitro antimicrobial activity of sulfonamides against some porcine pathogens. Am J Vet Res 50:1022–1028
Moore EJ, Kern EB (2001) Atrophic rhinitis: a review of 242 cases. Am J Rhinol 15:355–361
Nicholson TL, Brockmeier SL, Loving CL, Register KB, Kehrli ME Jr, Stibitz SE, Shore SM (2012) Phenotypic modulation of the virulent Bvg phase is not required for pathogenesis and transmission of Bordetella bronchiseptica in swine. Infect Immun 80:1025–1036
Nicholson TL, Brockmeier SL, Loving CL, Register KB, Kehrli ME Jr, Shore SM (2014) The Bordetella bronchiseptica type III secretion system is required for persistence and disease severity but not transmission in swine. Infect Immun 82:1092–1103
Niemann L, Müller P, Brauns J, Nathaus R, Schäkel F, Kipschull K, Höltig D, Wendt M, Schwarz S, Kadlec K (2018) Antimicrobial susceptibility and genetic relatedness of respiratory tract pathogens in weaner pigs over a 12-month period. Vet Microbiol 219:165–170
Park GY, Lee HM, Yu HJ, Son JS, Park SJ, Song KS (2018) Bordetella bronchiseptica bacteriophage suppresses B. bronchiseptica-induced inflammation in swine nasal turbinate cells. Genes Genomics 40:1383–1388
Pepin KM, Domsic J, McKenna R (2008) Genomic evolution in a virus under specific selection for host recognition. Infect Genet Evol 8:825–834
Petrovic A, Kostanjsek R, Rakhely G, Knezevic P (2017) The first Siphoviridae family bacteriophages infecting Bordetella bronchiseptica isolated from environment. Microb Ecol 73:368–377
Pósa R, Donkó T, Bogner P, Kovács M, Repa I, Magyar T (2011) Interaction of Bordetella bronchiseptica, Pasteurella multocida, and fumonisin B1 in the porcine respiratory tract as studied by computed tomography. Can J Vet Res 75:176–182
Prüller S, Rensch U, Meemken D, Kaspar H, Kopp PA, Klein G, Kehrenberg C (2015) Antimicrobial Susceptibility of Bordetella bronchiseptica isolates from Swine and companion animals and detection of resistance genes. PLoS One 10:e0135703
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Rees CE, Dodd CE (2006) Phage for rapid detection and control of bacterial pathogens in food. Adv Appl Microbiol 59:159–186
Vecht U, Wisselink HJ, van Dijk JE, Smith HE (1992) Virulence of Streptococcus suis type 2 strains in newborn germfree pigs depends on phenotype. Infect Immun 60:550–556
Acknowledgements
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) through the Technology Commercialization Support Program funded by the Ministry of Agriculture, Food, and Rural Affairs (MAFRA) (817029-03-2-SB010).
Author information
Authors and Affiliations
Contributions
GYP and KSS: designed study. GYP, HJY, SJP, and JSS: performed experiments. GYP, JSS, SJP, and HJC: analyzed data. GYP and KSS: wrote manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Ga Young Park, Hyun Jin Yu, Jee Soo Son, Sang Joon Park, Hee-Jae Cha, and Kyoung Seob Song declare that we have no conflict of interest.
Ethical approval
This article does not contain any studies with human subjects or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Park, G.Y., Yu, H.J., Son, J.S. et al. Specific bacteriophage of Bordetella bronchiseptica regulates B. bronchiseptica-induced microRNA expression profiles to decrease inflammation in swine nasal turbinate cells. Genes Genom 42, 441–447 (2020). https://doi.org/10.1007/s13258-019-00906-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-019-00906-7