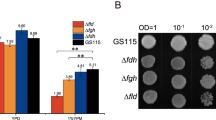
Abstract
Two catabolite repressor genes (MIG1 and MIG2) were previously identified in Pichia pastoris, and the derepression of alcohol oxidase (AOX) expression was realized in Δmig1 or Δmig1Δmig2 mutants grown in glycerol, but not in glucose. In this study, genome-wide RNA-seq analysis of Δmig1Δmig2 and the wild-type strain grown in glycerol revealed that the expression of numerous genes was greatly altered. Nearly 7% (357 genes) of approximately 5276 genes annotated in P. pastoris were significantly upregulated, with at least a two-fold differential expression in Δmig1Δmig2; the genes were mainly related to cell metabolism. Approximately 23% (1197 genes) were significantly downregulated; these were mainly correlated with the physiological characteristics of the cell. The methanol catabolism and peroxisome biogenesis pathways were remarkably enhanced, and the genes AOX1 and AOX2 were upregulated higher than 30-fold, which was consistent with the experimental results of AOX expression. The Mig proteins had a slight effect on autophagy when cells were grown in glycerol. The expression analysis of transcription factors showed that deletion of MIG1 and MIG2 significantly upregulated the binding of an essential transcription activator, Mit1p, with the AOX1 promoter, which suggested that Mig proteins might regulate the AOX1 promoter through the regulation of Mit1p. This work provides a reference for the further exploration of the methanol induction and catabolite repression mechanisms of AOX expression in methylotrophic yeasts.






Similar content being viewed by others
References
Alexa A, Rahnenführer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22:1600–1607
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B (Methodological) 57:289–300
Carlson M (1999) Glucose repression in yeast. Curr Opin Microbiol 2:202–207
Carmona TA, Barrado P, Jiménez A, Fernández Lobato M (2002) Molecular and functional analysis of a MIG1 homologue from the yeast Schwanniomyces occidentalis. Yeast 19:459–465
Cereghino JL, Cregg JM (2000) Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev 24:45–66
Chang T, Schroder LA, Thomson JM, Klocman AS, Tomasini AJ, Strømhaug PE, Dunn WA Jr (2005) Ppatg9 encodes a novel membrane protein that traffics to vacuolar membranes, which sequester peroxisomes during pexophagy in Pichia pastoris. Mol Biol Cell 16:4941–4953
De Schutter K, Lin YC, Tiels P, Van Hecke A, Glinka S, Weber-Lehmann J, Rouzé P, Van de Peer Y, Callewaert N (2009) Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol 27:561–566
Distel B, Van Der Ley I, Veenhuis M, Tabak HF (1988) Alcohol oxidase expressed under nonmethylotrophic conditions is imported, assembled, and enzymatically active in peroxisomes of Hansenula polymorpha. J Cell Biol 107:1669–1675
Egli T, Dijken JP, Veenhuis M, Harder W, Fiechter A (1980) Methanol metabolism in yeasts: regulation of the synthesis of catabolic enzymes. Arch Microbiol 124:115–121
Farré JC, Manjithaya R, Mathewson RD, Subramani S (2008) Ppatg30 tags peroxisomes for turnover by selective autophagy. Dev Cell 14:365–376
Gancedo JM (1998) Yeast carbon catabolite repression. Microbiol Mol Biol R 62:334–361
Gasser B, Prielhofer R, Marx H, Maurer M, Nocon J, Steiger M, Puxbaum V, Sauer M, Mattanovich D (2013) Pichia pastoris: protein production host and model organism for biomedical research. Future Microbiol 8:191–208
Hartner FS, Glieder A (2006) Regulation of methanol utilisation pathway genes in yeasts. Microb Cell Fact 5:39
Hazeu W, Bruyn JC, Bos P (1972) Methanol assimilation by yeasts. Arch Microbiol 87:185–188
Hedges D, Proft M, Entian KD (1995) CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol Cell Biol 15:1915–1922
Inan M, Meagher MM (2001) Non-repressing carbon sources for alcohol oxidase (AOX1) promoter of Pichia pastoris. J Biosci Bioeng 92:585–589
Kulmburg P, Mathieu M, Dowzer C, Kelly J, Felenbok B (1993) Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CREA repressor mediating carbon cataboiite repression in Aspergillus nidulans. Mol Microbiol 7:847–857
Leão-Helder AN, Krikken AM, Lunenborg MGJ, Kiel JA, Veenhuis M, van der Klei IJ (2004) Hansenula polymorpha Tup1p is important for peroxisome degradation. FEMS Yeast Res 4:789–794
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26:589–595
Li P, Anumanthan A, Gao XG, Ilangovan K, Suzara VV, Düzgüneş N, Renugopalakrishnan V (2007) Expression of recombinant proteins in Pichia pastoris. Appl Biochem Biotechnol 142:105–124
Lin-Cereghino GP, Godfrey L, de la Cruz BJ, Johnson S, Khuongsathiene S, Tolstorukov I, Yan M, Lin-Cereghino J, Veenhuis M, Subramani S, Cregg JM (2006) Mxr1p, a key regulator of the methanol utilization pathway and peroxisomal genes in Pichia pastoris. Mol Cell Biol 26:883–897
Marguerat S, Bähler J (2010) RNA-seq: from technology to biology. Cell Molr Life Sci 67:569–579
Mathieu M, Felenbok B (1994) The Aspergillus nidulans CREA protein mediates glucose repression of the ethanol regulon at various levels through competition with the ALCR-specific transactivator. EMBO J 13:4022–4027
Monastryska I, Kiel JAKW., Krikken AM, Komduur JA, Veenhuis M, van der Klei IJ (2005) The Hansenula polymorpha atg25 gene encodes a novel coiled-coil protein that is required for macropexophagy. Autophagy 1:92–100
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-SEq. Nat Methods 5:621–628
Nehlin JO, Ronne H (1990) Yeast MIG1 repressor is related to the mammalian early growth response and Wilms’ tumour finger proteins. EMBO J 9:2891–2898
Nehlin JO, Carlberg M, Ronne H (1991) Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J 10:3373–3377
Ostling J, Carlberg M, Ronne H (1996) Functional domains in the Mig1 repressor. Mol Cell Biol 16:753–761
Ozimek P, Veenhuis M, van der Klei IJ (2005) Alcohol oxidase: a complex peroxisomal, oligomeric flavoprotein. FEMS yeast Res 5:975–983
Ozsolak F, Milos PM (2010) RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12:87–98
Sahu U, Krishna Rao K, Rangarajan PN (2014) Trm1p, a Zn(II)2Cys6-type transcription factor, is essential for the transcriptional activation of genes of methanol utilization pathway, in Pichia pastoris. Biochem Biophys Res Commun 451:158–164
Sasano Y, Yurimoto H, Kuriyama M, Sakai Y (2010) Trm2p-dependent derepression is essential for methanol-specific gene activation in the methylotrophic yeast Candida boidinii. FEMS Yeast Res 10:535–544
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Schüller HJ (2003) Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr genet 43:139–160
Schultz J, Carlson M (1987) Molecular analysis of SSN6, a gene functionally related to the SNF1 protein kinase of Saccharomyces cerevisiae. Mol Cell Biol 7:3637–3645
Sibirny AA (2016) Yeast peroxisomes: structure, functions and biotechnological opportunities. FEMS Yeast Res 16:fow038
Stasyk OG, Van Zutphen T, Ah Kang H, Stasyk OV, Veenhuis M, Sibirny AA (2007) The role of Hansenula polymorpha MIG1 homologues in catabolite repression and pexophagy. FEMS Yeast Res 7:1103–1113
Stasyk OG, Maidan MM, Stasyk V, Van Dijck P, Thevelein JM, Sibirny AA (2008) Identification of hexose transporter-like sensor HXS1 and functional hexose transporter HXT1 in the methylotrophic yeast Hansenula polymorpha. Eukaryot cell 7:735–746
Van der Klei IJ, Yurimoto H, Sakai Y, Veenhuis M (2006) The significance of peroxisomes in methanol metabolism in methylotrophic yeast. BBA-Mol Cell Res 1763:1453–1462
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034
Vogl T, Glieder A (2013) Regulation of Pichia pastoris promoters and its consequences for protein production. New Biotechnol 30:385–404
Walther K, Schüller HJ (2001) Adr1 and Cat8 synergistically activate the glucose-regulated alcohol dehydrogenase gene ADH2 of the yeast. Microbiology 147:2037–2044
Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138
Wang X, Wang Q, Wang J, Bai P, Shi L, Shen W, Zhou M, Zhou X, Zhang Y, Cai M (2016) Mit1 transcription factor mediates methanol signaling and regulates alcohol oxidase 1 promoter in Pichia pastoris. J Biol Chem 291:6245–6261
Wang J, Wang X, Shi L, Qi F, Zhang P, Zhang Y, Zhou X, Song Z, Cai M (2017) Methanol-independent protein expression by AOX1 promoter with trans-acting elements engineering and glucose-glycerol-shift induction in Pichia pastoris. Sci Rep 7:41850
Wilhelm BT, Landry JR (2009) RNA-seq—quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48:249–257
Yurimoto H, Oku M, Sakai Y (2011) Yeast methylotrophy: metabolism, gene regulation and peroxisome homeostasis. Int J Microbiol 2011:101298
Zhang P, Zhang W, Zhou X, Bai P, Cregg JM, Zhang Y (2010) Catabolite repression of AOX in Pichia pastoris is dependent on hexose transporter PpHxt1 and pexophagy. Appl Environ Microbiol 76:6108–6118
Acknowledgements
This work was supported by Shanghai Science and Technology Innovation Plan (17JC1402400); Fundamental Research Funds for the Central Universities (22A201514040) and Grants of Young and Middle-aged Leading Science and Technology Innovation Talents from Ministry of Science and Technology of China. The authors are grateful to Prof. James M. Cregg, BioGrammatics, Inc. and Keck Graduate Institute of Applied Life Science, for helpful suggestions. The authors also thank Shanghai Huaguan Biochip Co. Ltd. for the help of RNA-seq data analysis.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Lei Shi declares that she has no conflict of interest. Xiaolong Wang declares that he has no conflict of interest. Jinjia Wang declares that she has no conflict of interest. Ping Zhang declares that she has no conflict of interest. Fei Qi declares that he has no conflict of interest. Menghao Cai declares that he has no conflict of interest. Yuanxing Zhang declares that he has no conflict of interest. Xiangshan Zhou declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shi, L., Wang, X., Wang, J. et al. Transcriptome analysis of Δmig1Δmig2 mutant reveals their roles in methanol catabolism, peroxisome biogenesis and autophagy in methylotrophic yeast Pichia pastoris . Genes Genom 40, 399–412 (2018). https://doi.org/10.1007/s13258-017-0641-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-017-0641-5