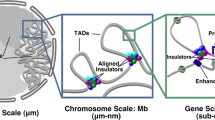
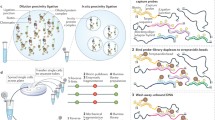
Abstract
The nucleus of every cell is arranged in a manner that reflects the cell’s functional identity. Its DNA loops around histones that help compact and regulate the local functional state. Various other proteins along with RNAs are arranged along the DNA. Barrier and insulator elements partition it into functional elements. Other proteins interact with their binding sites, recruit other factors, to enable the modifying of histones according to the functional state it must be in. Depending on this state, other proteins can facilitate transcription, repair, replication etc. To this end, the study of the spatial distribution of these proteins and RNAs is necessary to understand their functional relevance within the context of the nucleus. Here, we review various established and emerging techniques that enable such studies.




Similar content being viewed by others
References
Ashburner M, Roote J. Maintenance of a Drosophila laboratory: general procedures. CSH Protoc. 2007. https://doi.org/10.1101/pdb.ip35.
Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Inductions of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med. 1944;79(2):137–58.
Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300(5620):764. https://doi.org/10.1126/science.1079512.
Bohn M, Diesinger P, Kaufmann R, Weiland Y, Müller P, Gunkel M, et al. Localization microscopy reveals expression-dependent parameters of chromatin nanostructure. Biophys J. 2010;99(5):1358–67. https://doi.org/10.1016/j.bpj.2010.05.043.
Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.9.1-9. https://doi.org/10.1002/0471142727.mb2129s109.
Byrum SD, Raman A, Taverna SD, Tackett AJ. ChAP-MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell Rep. 2012;2(1):198–205. https://doi.org/10.1016/j.celrep.2012.06.019.
Cerase A, Pintacuda G, Tattermusch A, Avner P. Xist localization and function: new insights from multiple levels. Genome Biol. 2015;16:166. https://doi.org/10.1186/s13059-015-0733-y.
Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155(7):1479–91. https://doi.org/10.1016/j.cell.2013.12.001.
Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA–chromatin interactions. Mol Cell. 2011;44(4):667–78. https://doi.org/10.1016/j.molcel.2011.08.027.
Cremer M, Grasser F, Lanctôt C, Müller S, Neusser M, Zinner R, et al. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol Biol. 2008;463:205–39. https://doi.org/10.1007/978-1-59745-406-3_15.
Dambournet D, Hong SH, Grassart A, Drubin DG. Tagging endogenous loci for live-cell fluorescence imaging and molecule counting using ZFNs, TALENs, and Cas9. Methods Enzymol. 2014;546:139–60. https://doi.org/10.1016/B978-0-12-801185-0.00007-6.
Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136(1):175–86. https://doi.org/10.1016/j.cell.2008.11.045.
Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–11. https://doi.org/10.1126/science.1067799.
Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518(7539):331–6. https://doi.org/10.1038/nature14222.
Doncaster L. The mechanism of Mendelian heredity. Eugen Rev 1916;8(2):164–166.
Dorn R, Szidonya J, Korge G, Sehnert M, Taubert H, Archoukieh E, et al. P transposon-induced dominant enhancer mutations of position-effect variegation in Drosophila melanogaster. Genetics. 1993;133(2):279–90.
Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16(10):1299–309. https://doi.org/10.1101/gr.5571506.
Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–5. https://doi.org/10.1038/nature20149.
Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. https://doi.org/10.1126/science.1237973.
Fabre PJ, Leleu M, Mormann BH, Lopez-Delisle L, Noordermeer D, Beccari L, et al. Large scale genomic reorganization of topological domains at the HoxD locus. Genome Biol. 2017;18(1):149. https://doi.org/10.1186/s13059-017-1278-z.
Feng S-Y, Ota K, Yamada Y, Sawabu N, Ito T. A yeast one-hybrid system to detect methylation-dependent DNA–protein interactions. Biochem Biophys Res Commun. 2004;313(4):922–5. https://doi.org/10.1016/j.bbrc.2003.12.027.
Franklin RE, Gosling RG. Molecular configuration in sodium thymonucleate. Nature. 1953;171(4356):740–1. https://doi.org/10.1038/171740a0.
Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462(7269):58–64. https://doi.org/10.1038/nature08497.
Goryshin IY, Reznikoff WS. Tn5 in vitro transposition. J Biol Chem. 1998;273(13):7367–74. https://doi.org/10.1074/jbc.273.13.7367.
Gustafsson MGL. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. Short communication. J Microsc. 2000;198(2):82–7. https://doi.org/10.1046/j.1365-2818.2000.00710.x.
Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21(2):198–206. https://doi.org/10.1038/nsmb.2764.
Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19(11):780–2. https://doi.org/10.1364/OL.19.000780.
Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91(11):4258–72. https://doi.org/10.1529/biophysj.106.091116.
Jackson DA, Iborra FJ, Manders EM, Cook PR. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol Biol Cell. 1998;9(6):1523–36.
James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6(11):3862–72.
Jäger R, Migliorini G, Henrion M, Kandaswamy R, Speedy HE, Heindl A, et al. Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat Commun. 2015;6:6178. https://doi.org/10.1038/ncomms7178.
Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell. 2015;163(1):134–47. https://doi.org/10.1016/j.cell.2015.08.040.
Kohalmi SE, Kunz BA. Role of neighbouring bases and assessment of strand specificity in ethylmethanesulphonate and N-methyl-N′-nitro-N-nitrosoguanidine mutagenesis in the SUP4-o gene of Saccharomyces cerevisiae. J Mol Biol. 1988;204(3):561–8. https://doi.org/10.1016/0022-2836(88)90355-5.
Koohy H, Down TA, Hubbard TJ. Chromatin accessibility data sets show bias due to sequence specificity of the DNase I enzyme. PLoS ONE. 2013;8(7):e69853. https://doi.org/10.1371/journal.pone.0069853.
Langer-Safer PR, Levine M, Ward DC. Immunological method for mapping genes on Drosophila polytene chromosomes. Proc Natl Acad Sci USA. 1982;79(14):4381–5.
Larsen BD, Madsen MR, Nielsen R, Mandrup S. Chromatin immunoprecipitation for identification of protein–DNA interactions in human cells. Methods Mol Biol. 2018;1794:335–52. https://doi.org/10.1007/978-1-4939-7871-7_24.
Lewis EB. The phenomenon of position effect. Adv Genet 1950;3:73–115. https://doi.org/10.1016/S0065-2660(08)60083-8
Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat Genet. 2001;28(4):327–34. https://doi.org/10.1038/ng569.
Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–93. https://doi.org/10.1126/science.1181369.
Liu X, Zhang Y, Chen Y, Li M, Shao Z, Zhang MQ, et al. CAPTURE: in situ analysis of chromatin composition of endogenous genomic loci by biotinylated dCas9. Curr Protoc Mol Biol. 2018. https://doi.org/10.1002/cpmb.64.
Luo S, Lu J. Silencing of transposable elements by piRNAs in Drosophila: an evolutionary perspective. Genom Proteom Bioinform. 2017;15(3):164–76. https://doi.org/10.1016/j.gpb.2017.01.006.
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. https://doi.org/10.1126/science.1232033.
Mavor JW. An effect of X rays on the linkage of mendelian characters in the first chromosome of Drosophila. Genetics. 1923;8(4):355–66.
Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–8. https://doi.org/10.1038/nbt.1755.
Muller HJ. Types of visible variations induced by X-rays in Drosophila. J Genet. 1930;22(3):299–334. https://doi.org/10.1007/BF02984195.
Nagano T, Lubling Y, Yaffe E, Wingett SW, Dean W, Tanay A, et al. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat Protoc. 2015;10(12):1986–2003. https://doi.org/10.1038/nprot.2015.127.
Nowoshilow S, Schloissnig S, Fei J-F, Dahl A, Pang AWC, Pippel M, et al. The axolotl genome and the evolution of key tissue formation regulators. Nature. 2018;554(7690):50–5. https://doi.org/10.1038/nature25458.
Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120(3):621–3.
O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–45. https://doi.org/10.1093/nar/gkv1189.
Pennisi E. ENCODE project writes eulogy for junk DNA. Science. 2012;337(6099):1159–61. https://doi.org/10.1126/science.337.6099.1159.
Podlevsky JD, Chen JJL. Evolutionary perspectives of telomerase RNA structure and function. RNA Biol. 2016;13(8):720–32. https://doi.org/10.1080/15476286.2016.1205768.
Port F, Chen H-M, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 2014;111(29):E2967–76. https://doi.org/10.1073/pnas.1405500111.
Reuter G, Dorn R, Wustmann G, Friede B, Rauh G. Third chromosome suppressor of position-effect variegation loci in Drosophila melanogaster. Mol Gen Genet. 1986;202(3):481–7. https://doi.org/10.1007/BF00333281.
Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4(8):651–7. https://doi.org/10.1038/nmeth1068.
Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33(11):1165–72. https://doi.org/10.1038/nbt.3383.
Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20(6):761–70. https://doi.org/10.1101/gr.099655.109.
Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320(5881):1332–6. https://doi.org/10.1126/science.1156947.
Schmitt AD, Hu M, Ren B. Genome-wide mapping and analysis of chromosome architecture. Nat Rev Mol Cell Biol. 2016;17(12):743–55. https://doi.org/10.1038/nrm.2016.104.
Shahi P, Kim SC, Haliburton JR, Gartner ZJ, Abate AR. Abseq: ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding. Sci Rep. 2017;7:44447. https://doi.org/10.1038/srep44447.
Simon JM, Giresi PG, Davis IJ, Lieb JD. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc. 2012;7(2):256–67. https://doi.org/10.1038/nprot.2011.444.
Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504(7480):465–9. https://doi.org/10.1038/nature12719.
Sinha RP, Häder DP. UV-induced DNA damage and repair: a review. Photochem Photobiol Sci. 2002;1(4):225–36. https://doi.org/10.1039/b201230h.
Song F, Chen P, Sun D, Wang M, Dong L, Liang D, et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344(6182):376–80. https://doi.org/10.1126/science.1251413.
Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010. https://doi.org/10.1101/pdb.prot5384.
Srivastava S, Dhawan J, Mishra RK. Epigenetic mechanisms and boundaries in the regulation of mammalian Hox clusters. Mech Dev. 2015;138(Pt 2):160–9. https://doi.org/10.1016/j.mod.2015.07.015.
Srivastava S, Puri D, Garapati HS, Dhawan J, Mishra RK. Vertebrate GAGA factor associated insulator elements demarcate homeotic genes in the HOX clusters. Epigenetics Chromatin. 2013;6(1):8. https://doi.org/10.1186/1756-8935-6-8.
Srivastava S, Sowpati DT, Garapati HS, Puri D, Dhawan J, Mishra RK. A ChIP-on-chip tiling array approach detects functional histone-free regions associated with boundaries at vertebrate HOX genes. Genom Data. 2014;2:78–81. https://doi.org/10.1016/j.gdata.2014.05.001.
van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18(4):424–8. https://doi.org/10.1038/74487.
Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36(3):283–7. https://doi.org/10.1038/ng1314.
Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–46. https://doi.org/10.1038/nrg2842.
Wang X, Fang X, Yang P, Jiang X, Jiang F, Zhao D, et al. The locust genome provides insight into swarm formation and long-distance flight. Nat Commun. 2014;5:2957. https://doi.org/10.1038/ncomms3957.
Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–8. https://doi.org/10.1038/171737a0.
Wen L, Tang F. Single cell epigenome sequencing technologies. Mol Aspects Med. 2018;59:62–9. https://doi.org/10.1016/j.mam.2017.09.002.
Wilkins MHF, Stokes AR, Wilson HR. Molecular structure of deoxypentose nucleic acids. Nature. 1953;171(4356):738–40.
Woodcock CLF, Safer JP, Stanchfield JE. Structural repeating units in chromatin. Exp Cell Res. 1976;97(1):101–10. https://doi.org/10.1016/0014-4827(76)90659-5.
Wu X-S, Wang F, Li H-F, Hu Y-P, Jiang L, Zhang F, et al. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 2017;18(10):1837–53. https://doi.org/10.15252/embr.201744147.
Zaret K. Micrococcal nuclease analysis of chromatin structure. Curr Protoc Mol Biol. 2005. https://doi.org/10.1002/0471142727.mb2101s69.
Zessin PJM, Finan K, Heilemann M. Super-resolution fluorescence imaging of chromosomal DNA. J Struct Biol. 2012;177(2):344–8. https://doi.org/10.1016/j.jsb.2011.12.015.
Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38(11):1341–7. https://doi.org/10.1038/ng1891.
Acknowledgements
TP acknowledges the Council of Scientific and Industrial Research (CSIR) for doctoral fellowship and RKM acknowledges the financial support of the CSIR.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is dedicated in memory of Profs AK Sharma and Archana Sharma.
Rights and permissions
About this article
Cite this article
Ponrathnam, T., Mishra, R.K. From chromosomes to genomes: new insights with emerging techniques. Nucleus 61, 227–234 (2018). https://doi.org/10.1007/s13237-018-0242-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-018-0242-y