Abstract
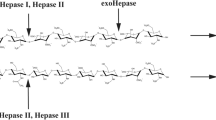
Heparinases are widely used for production of clinically and therapeutically important bioactive oligosaccharides and in analyzing the polydisperse, heterogeneous, and complex structures of heparin/heparan sulfate. In the present study, the gene (1911 bp) encoding heparinase II/III of family 12 polysaccharide lyase (PsPL12a) from Pseudopedobacter saltans was cloned, expressed, and biochemically and functionally characterized. The purified enzyme PsPL12a of molecular size approximately 76 kDa exhibited maximum activity in the temperature range 45–50 °C and at pH 6.0. PsPL12a gave maximum activity at 1% (w/v) heparin under optimum conditions. The kinetic parameters, K m and Vmax, for PsPL12a were 4.6 ± 0.5 mg/ml and 70 ± 2 U/mg, respectively. Ten millimolars of each Mg2+ and Mn2+ ions enhanced PsPL12a activity by 80%, whereas Ni2+ inhibited by 75% and Co2+ by 10%, and EDTA completely inactivated the enzyme. Protein melting curve of PsPL12a gave a single peak at 55 °C and 10 mM Mg2+ ions and shifted the peak to 60 °C. The secondary structure analysis of PsPL12a by CD showed 65.12% α-helix, 11.84% β-strand, and 23.04% random coil. The degradation products of heparin by PsPL12a analyzed by ESI-MS spectra displayed peaks corresponding to heparin di-, tetra-, penta-, and hexa-saccharides revealing the endolytic mode of enzyme action. Heparinase II/III (PsPL12a) from P. saltans can be used for production of low molecular weight heparin oligosaccharides for their utilization as anticoagulants. This is the first report on heparinase cloned from P. saltans.







Similar content being viewed by others
References
Amin K (2012) The role of mast cells in allergic inflammation. Respir Med 106(1):9–14
Bellamy RW, Horikoshi K (1992) U.S. patent no. 5,145,778. U.S. Patent and Trademark Office, Washington, DC
Böhmer LH, Pitout MJ, Steyn PL, Visser L (1990) Purification and characterization of a novel heparinase. J Biol Chem 265(23):13609–13617
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1–2):248–254
Cao J, Lai Q, Li G, Shao Z (2014) Pseudopedobacter beijingensis gen. nov., sp. nov., isolated from coking wastewater activated sludge, and reclassification of Pedobacter saltans as Pseudopedobacter saltans comb. nov. Int J Syst Evol Microbiol 64(6):1853–1858
Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, Marks RM (1997) Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med 3(8):866–871
Chen S, Ye F, Chen Y, Chen Y, Zhao H, Yatsunami R, Nakamura S, Arisaka F, Xing XH (2011) Biochemical analysis and kinetic modeling of the thermal inactivation of MBP-fused heparinase I: implications for a comprehensive thermostabilization strategy. Biotechnol Bioeng 108(8):1841–1851
Davies G, Henrissat B (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3(9):853–859
Desai UR, Wang HM, Linhardt RJ (1993) Specificity studies on the heparin lyases from Flavobacterium heparinum. Biochemistry 32(32):8140–8145
Doneanu CE, Chen W, Gebler JC (2009) Analysis of oligosaccharides derived from heparin by ion-pair reversed-phase chromatography/mass spectrometry. Anal Chem 81(9):3485–3499
Dreyfuss JL, Regatieri CV, Jarrouge TR, Cavalheiro RP, Sampaio LO, Nader HB (2009) Heparan sulfate proteoglycans: structure, protein interactions and cell signaling. An Acad Bras Cienc 81(3):409–429
Ernst S, Langer R, Cooney CL, Sasisekharan R (1995) Enzymatic degradation of glycosaminogIycans. Crit Rev Biochem Mol Biol 30(5):387–444
Esko JD, Selleck SB (2002) Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem 71(1):435–471
Gallagher JT, Turnbull JE (1992) Heparan sulphate in the binding and activation of basic fibroblast growth factor. Glycobiology 2(6):523–528
Garron ML, Cygler M (2010) Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 20(12):1547–1573
Gasimli L, Robert JL, Dordick JS (2012) Proteoglycans in stem cells. Biotechnol Appl Biochem 59(2):65–76
Gray E, Hogwood J, Mulloy B (2012) The anticoagulant and antithrombotic mechanisms of heparin. In: Heparin-A century of Progress. Springer, Berlin Heidelberg, pp 43–61
Hovingh P, Linker A (1970) The enzymatic degradation of heparin and heparitin sulfate III. Purification of a heparitinase and a heparinase from flavobacteria. J Biol Chem 245(22):6170–6175
Hulett MD, Hornby JR, Ohms SJ, Zuegg J, Freeman C, Gready JE, Parish CR (2000) Identification of active-site residues of the pro-metastatic endoglycosidase heparanase. Biochemistry 39(51):15659–15667
Hyun YJ, Lee JH, Kim DH (2010) Cloning, overexpression, and characterization of recombinant heparinase III from Bacteroides stercoris HJ-15. Appl Microbiol Biotechnol 86(3):879–890
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Limtiaco JF, Beni S, Jones CJ, Langeslay DJ, Larive CK (2011) NMR methods to monitor the enzymatic depolymerization of heparin. Anal Bioanal Chem 399(2):593–603
Lindahl U, Lidholt K, Spillmann D, Kjellén L (1994) More to “heparin” than anticoagulation. Thromb Res 75(1):1–32
Linhardt RJ, Fitzgerald GL, Cooney CL, Langer R (1982) Mode of action of heparin lyase on heparin. Biochim Biophys Acta Protein Struct Mol Enzymol 702(2):197–203
Linhardt RJ, Turnbull JE, Wang HM, Loganathan D, Gallagher JT (1990) Examination of the substrate specificity of heparin and heparan sulfate lyases. Biochemistry 29(10):2611–2617
Lohse DL, Linhardt RJ (1992) Purification and characterization of heparin lyases from Flavobacterium heparinum. J Biol Chem 267(34):24347–24355
Lundin L, Larsson H, Kreuger J, Kanda S, Lindahl U, Salmivirta M, Claesson-Welsh L (2000) Selectively desulfated heparin inhibits fibroblast growth factor-induced mitogenicity and angiogenesis. J Biol Chem 275(32):24653–24660
Nader HB, Porcionatto MA, Tersariol IL, Pinhal MA, Oliveira FW, Moraes CT, Dietrich CP (1990) Purification and substrate specificity of heparitinase I and heparitinase II from Flavobacterium heparinum. Analyses of the heparin and heparan sulfate degradation products by 13C NMR spectroscopy. J Biol Chem 265(28):16807–16813
Olsson P, Sanchez J, Mollnes TE, Riesenfeld J (2000) On the blood compatibility of end-point immobilized heparin. J Biomater Sci Polym Ed 11(11):1261–1273
Rabenstein DL (2002) Heparin and heparan sulfate: structure and function. Nat Prod Rep 19(3):312–331
Saad OM, Ebel H, Uchimura K, Rosen SD, Bertozzi CR, Leary JA (2005) Compositional profiling of heparin/heparan sulfate using mass spectrometry: assay for specificity of a novel extracellular human endosulfatase. Glycobiology 15(8):818–826
Sampaio LO, Tersariol IL, Lopes CC, Bouças RI, Nascimento FD, Rocha HA, Nader HB (2006) Heparins and heparans sulfates. Structure, distribution and protein interactions. Insights into Carbohydrate Structure and Bological Function, 1–24
Sarrazin S, Lamanna WC, Esko JD (2011) Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol 3(7):a004952
Shively JE, Conrad HE (1976) Formation of anhydrosugars in the chemical depolymerization of heparin. Biochemistry 15(18):3932–3942
Shriver Z, Capila I, Venkataraman G, Sasisekharan R (2012) Heparin and heparan sulfate: analyzing structure and microheterogeneity. Heparin-A Century of Progress. Springer Berlin Heidelberg. 159–176
Steyn PL, Segers P, Vancanneyt M, Sandra P, Kersters K, Joubert JJ (1998) Classification of heparinolytic bacteria into a new genus, Pedobacter, comprising four species: Pedobacter heparinus comb. nov., Pedobacter piscium comb. nov., Pedobacter africanus sp. nov. and Pedobacter saltans sp. nov. proposal of the family Sphingobacteriaceae fam. nov. Int J Syst Evol Microbiol 48(1):165–177
Thanawiroon C, Rice KG, Toida T, Linhardt RJ (2004) Liquid chromatography/mass spectrometry sequencing approach for highly sulfated heparin-derived oligosaccharides. J Biol Chem 279(4):2608–2615
Toyoshima M, Nakajima M (1999) Human heparanase purification, characterization, cloning, and expression. J Biol Chem 274(34):24153–24160
Xiao Z, Zhao W, Yang B, Zhang Z, Guan H, Linhardt RJ (2010) Heparinase 1 selectivity for the 3, 6-di-O-sulfo-2-deoxy-2-sulfamido-α-D-glucopyranose (1, 4) 2-O-sulfo-α-L-idopyranosyluronic acid (GlcNS3S6S-IdoA2S) linkages. Glycobiology 21(1):13–22
Yamada S, Sugahara K (2003) Preparation of oligosaccharides from sulfated glycosaminoglycans using bacterial enzymes. In: Capillary electrophoresis of carbohydrates, pp 71–78
Yates EA, Rudd TR (2016) Recent innovations in the structural analysis of heparin. Int J Cardiol 212:S5–S9
Yoshida E, Sakai K, Tokuyama S, Miyazono H, Maruyama H, Morikawa K, Tahara Y (2002) Purification and characterization of heparinase that degrades both heparin and heparan sulfate from Bacillus circulans. Biosci Biotechnol Biochem 66(5):1181–1184
Acknowledgements
The authors thank DBT Program Support, IIT Guwahati, for CD analysis and Central Instrumentation Facility for ESI-mass analysis. Fellowship provided by the Ministry of Human Resource Development, Govt. of India, to Karthika B. is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Balasubramaniam, K., Sharma, K., Rani, A. et al. Deciphering the mode of action, structural and biochemical analysis of heparinase II/III (PsPL12a) a new member of family 12 polysaccharide lyase from Pseudopedobacter saltans. Ann Microbiol 68, 409–418 (2018). https://doi.org/10.1007/s13213-018-1347-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-018-1347-x