Abstract
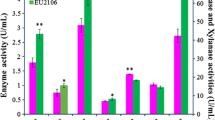
A transfer DNA (T-DNA)-tagged mutant library of Humicola insolens was screened for mutants with altered cellulase production using the plate-clearing zone assay. Three selected mutants (5-A7, 5-C6, and 13-B7) exhibited significantly depressed FPase, CMCase and xylanase activities compared with the wild-type strain upon shake-flask fermentation, while the pNPCase and pNPGase activities of the three mutants were relatively higher than those of the parental strain. Combined with the results of SDS-PAGE and mass spectrometry, we suggest that expression of the CMCases Cel6B, Cel7B, CMC3, and XynA/B/C was reduced in the mutant strains. Twelve putative T-DNA insertion sites were identified in the three mutants via Agrobacterium-mediated insertional mutagenesis sequencing (AIM-Seq). Bioinformatics analysis suggested that a putative dolichyl pyrophosphate phosphatase, two hypothetical proteins encoding genes of unknown function, and/or nine intergenic fragments may be involved in cellulase and hemicellulase production by H. insolens. This provides promising new candidate genes relevant to cellulase production by the fungus, which will be crucial not only for our understanding of the molecular mechanism underlying cellulase production, but also for strain improvement.



Similar content being viewed by others
References
Chacko N, Zhao Y, Yang E, Wang L, Cai JJ, Lin X (2015) The lncRNA RZE1 Controls Cryptococcal morphological transition. PLoS Genet 11:e1005692
Dalbøge H, HeldtHansen HP (1994) A novel method for efficient expression cloning of fungal enzyme genes. Mol Gen Genet 243:253–260
Davies GJ, Ducros V, Lewis RJ, Borchert TV, Schülein M (1997) Oligosaccharide specificity of a family 7 CMCase: insertion of potential sugar-binding subsites. J Biotechnol 57:91–100
Davies GJ, Brzozowski AM, Dauter M, Varrot A, Schulein M (2000) Structure and function of Humicola insolens family 6 cellulases: structure of the CMCase, Cel6B, at 1.6 A resolution. Biochem J 348:201–207
Du Y, Shi P, Huang H, Zhang X, Luo H, Wang Y, Yao B (2013) Characterization of three novel thermophilic xylanases from Humicola insolens Y1 with application potentials in the brewing industry. Bioresour Technol 130:161–167
Esher SK, Granek JA, Alspaugh JA (2015) Rapid mapping of insertional mutations to probe cell wall regulation in Cryptococcus neoformans. Fungal Genet Biol 82:9–21
James AW, Gowsalya R, Nachiappan V (2016) Dolichyl pyrophosphate phosphatase-mediated N-glycosylation defect dysregulates lipid homeostasis in Saccharomyces cerevisiae. Biochim Biophys Acta 1861:1705–1718
Kemski MM, Stevens B, Rappleye CA (2013) Spectrum of T-DNA integrations for insertional mutagenesis of Histoplasma capsulatum. Fungal Biol 117:41–51
Le Nours J, Ryttersgaard C, Lo Leggio L, Østergaard PR, Borchert TV, Christensen LLH, Larsen S (2003) Structure of two fungal β-1,4-galactanases: searching for the basis for temperature and pH optimum. Protein Sci 12:1195–1204
Li G, Zhou Z, Liu G, Zheng F, He C (2007) Characterization of T-DNA insertion patterns in the genome of rice blast fungus Magnaporthe oryzae. Curr Genet 51:233–243
Long L, Wang Y, Yang J, Xu X, Liu G (2013) A septation related gene AcsepH in Acremonium chrysogenum is involved in the cellular differentiation and cephalosporin production. Fungal Genet Biol 50:11–20
Meleiro LP, Zimbardi AL, Souza FH, Masui DC, Silva TM, Jorge JA, Furriel RP (2014) A novel pNPGase from Humicola insolens with high potential for untreated waste paper conversion to sugars. Appl Biochem Biotechnol 173:391–408
Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF (2005) Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr Genet 48:1–17
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Mullins ED, Chen X, Romaine P, Raina R, Geiser DM, Kang S (2001) Agrobacterium-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91:173–180. https://doi.org/10.1094/phyto.2001.91.2.173
Schulein M (1997) Enzymatic properties of cellulases from Humicola insolens. J Biotechnol 57:71–81
Tzfira T, Li J, Lacroix B, Citovsky V (2004) Agrobacterium T-DNA integration: molecules and models. Trends Genet Tig 20:375
Varrot Annabelle, Schülein Martin, Davies Gideon J (1999) Structural changes of the active site tunnel of Humicola insolens cellobiohydrolase Cel6A upon oligosaccharide binding. Biochemistry US 38:8884–8891
Xu X, Li J, Shi P, Ji W, Liu B, Zhang Y, Yao B, Fan L, Zhang W (2016) The use of T-DNA insertional mutagenesis to improve cellulase production by the thermophilic fungus Humicola insolens Y1. Sci Rep UK 6:31108
Yang X, Shi P, Huang H, Luo H, Wang Y, Zhang W, Yao B (2014) Two xylose-tolerant GH43 bifunctional β-xylosidase/α-arabinosidases and one GH11 xylanase from Humicola insolens and their synergy in the degradation of xylan. Food Chem 148:381–387
Zhong Y, Wang X, Yu H, Liang S, Wang T (2012) Application of T-DNA insertional mutagenesis for improving cellulase production in the filamentous fungus Trichoderma reesei. Bioresour Technol 110:572–577
Acknowledgements
This research was supported by the National Natural Science Foundation of China (Grant no. 31600065) and the Open Research Fund Program of the Beijing Key Laboratory of Plant Resource Research and Development, Beijing Technology and Business University (Grant no. 2016-YB3).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: XX, CF, PS, and WZ. Performed the experiments: CF, XX, and JL. Analyzed the data: CF, XX, LS, WG, and BL. Contributed reagents/materials/analysis tools: WZ. Wrote the paper: XX and CF.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fan, C., Xu, X., Song, L. et al. The use of Agrobacterium-mediated insertional mutagenesis sequencing to identify novel genes of Humicola insolens involved in cellulase production. 3 Biotech 8, 153 (2018). https://doi.org/10.1007/s13205-018-1166-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-018-1166-6