Abstract
Lactic acid bacteria, Enterococcus faecium and Lactococcus lactis, previously isolated from Thai fermented sausages were elucidated their probiotic properties especially in the control of Clostridium difficile 630. Both isolates survived in simulated gastric solution at pH 3 followed in simulated intestinal solution at pH 8. The presence of skimmed milk also helped the bacteria to survive through acidic and alkaline in gastrointestinal conditions. The adhesion properties of both isolates were tested using a human colon adenocarcinoma cell line. The result showed that both isolates exhibited desirable probiotic properties which adhered to Caco-2 cells. The neutralized cell-free supernatant of both isolates demonstrated that no cytotoxicity toward Caco-2 cells vice versa cell-free supernatant of C. difficile 630 toward Caco-2 cell demonstrated high toxicity. The immunomodulation effect in response to bacterial neutralized cell-free supernatant and cell-free supernatant was also studied. The expression level of pro-inflammatory cytokine of Caco-2 cell which are tumor necrosis factor-α and interleukin-8 was evaluated using quantitative reverse transcriptase PCR. Both isolates were able to diminish the expression level of TNF-α and IL-8 induced by the cell-free supernatant of C. difficile 630. Hence, these isolates would be able to improve the gut health through counteracting the C. difficile-associated intestinal inflammation in human cell lines. These results may contribute to the development of the isolates using as probiotics.
Similar content being viewed by others
Introduction
Lactic acid bacteria (LAB) have been widely used in the food manufacturing industry, primarily for fermentation processes. They are now frequently used as probiotics, bacteria that promote better health and well-being. Their proposed mode of action is referred to as colonization resistance, the creation of a physical barrier which enhances the integrity of tight junctions, and preventing the entry of pathogens. They possess antimicrobial activity via the production of antimicrobial peptides known as bacteriocins. They can also stimulate the production of anti-inflammatory cytokines, and inhibit or trigger an innate immune response by initiating TNF-α production to recruit neutrophils to the site of infection [1, 2].
Clostridium difficile is a Gram-positive, spore-forming anaerobic bacterium. It is one of the major causes of nosocomial diarrhea, ranging from mild diarrhea to life-threatening pseudomembranous colitis. The organism produces two major virulence toxins known as C. difficile toxin A (TcdA) and C. difficile toxin B (TcdB) that play crucial roles in the disruption of the actin cytoskeleton and the impairment of tight junctions in the intestinal epithelial cells, resulting in extensive damage to the large intestine [3,4,5]. C. difficile is thought to be a multidrug-resistant pathogen; due to its broad-spectrum antibiotic resistance, different alternative methodologies—including the use of probiotic bacteria such as LAB—have been investigated for the prevention and treatment of C. difficile infection (CDI).
Previous studies have reported the effects of LAB against C. difficile. Lactic acid produced by Streptococcus thermophilus inhibited the growth of C. difficile in a dose-dependent manner and decreased gene expression of TdcA [6]. Another study demonstrated that Lactobacillus rhamnosus GG displayed inhibitory effects on C. difficile growth and protected kidney epithelial cells from African green monkey (Vero cells) from toxigenic C. difficile [7]. Various strains of LAB such as L. rhamnosus LR5, Lactococcus lactis SL3, Bifidobacterium breve BR3, and Bifidobacterium lactis BL3 have demonstrated antimicrobial activity by inhibiting the growth of C. difficile [8].
In the present study, we aim to elucidate the probiotic properties of E. faecium and L. lactis isolated from Thai fermented sausages that demonstrated antimicrobial activity via the production of bacteriocins [9]. The properties include the ability to survive in simulated gastrointestinal solutions, adhesive properties to epithelium cell, cytotoxicity, and immunomodulation against multi-drug resistant C. difficile strain 630 (CD630) which is PCR-ribotype 012, PCR ribotyping of clinically important hypervirulent strain of C. difficile. The results will provide essential information regarding the use of these bacteria as probiotics.
Materials and Methods
Strains and Bacterial Culture Conditions
E. faecium and L. lactis producing bacteriocins, such as enterocin and nisin, respectively, were isolated from Thai fermented sausages [9]. Briefly, Thai fermented sausage samples were bought from local market, and samples were mixed with Criterion™ Lactobacilli de Man, Rogosa, and Sharpe (MRS) broth (Hardy Diagnostics, Santa Maria, CA, USA). After incubation at 37 °C for 1 h with continuous shaking, the samples were tenfold serially diluted and the appropriated dilutions were then spread onto MRS agar. The plate was then incubated at 37 °C for 24 h. The strains were identified by determination of 16S ribosomal RNA gene using universal primers for 16S rDNA of bacteria [10].
The bacteria were maintained on MRS agar at 37 °C for 24 h. The single colony of LAB was inoculated into MRS broth and further incubated at 37 °C for 24 h under aerobic condition. CD630, multi-drug resistant strain, was kindly provided by Prof. Nigel Minton, University of Nottingham, UK, and was streaked on Brain Heart Infusion (BHI) agar (Himedia, Mumbai, India) and incubated anaerobically at 37 °C for 24 h to obtain single colonies. BHI broth supplemented with 0.1% sodium taurocholate (TCI, Chuo-ku, Tokyo, Japan) and 0.5% yeast extract (BD, Franklin Lakes, NJ, USA) was inoculated with single colonies of CD630 and incubated under anaerobic condition for use in subsequent tests. For long-term storage, overnight cultures of each strain were transferred into sterilized glycerol to obtain a final concentration of 20% and kept at − 80 °C.
Preparation of the NCFS of LAB and CFS of CD630
Fifty milliliters of MRS broth was inoculated with 1% (v/v) of E. faecium and L. lactis culture as described above. The cultures were incubated at 37 °C for 16–18 h under aerobic condition. Bacterial supernatant was collected after centrifugation at 8000×g at 4 °C for 10 min and were neutralized using 1 M NaOH following by filtration through a sterilized 0.22-μm filter (Merck, Kenilworth, NJ, USA) to obtain neutralized cell-free supernatant (NCFS). CD630 was cultivated in BHI broth as mentioned earlier. Fifty milliliters of BHI broth was inoculated with 1% (v/v) of CD630 culture and further incubated at 37 °C for 24 h anaerobically. CD630 supernatant was also collected and filtered as described previously to obtain cell-free supernatant (CFS).
Total protein of the NCFS of E. faecium and L. lactis isolates and the CFS of CD630 were measured using Bradford solution (Bio-Rad, Hercules, CA, USA) following the manufacturer’s protocol. Bovine serum albumin (Takara, Kusatsu-ku, Shiga, Japan) was used as a standard protein. The optical density (OD) was measured at λ = 595 nm using UV1800 Spectrophotometer (Shimadzu, Nakagyo-ku, Kyoto, Japan).
Assay of Probiotics Properties
Survival of LAB in Simulated Gastrointestinal Solution
The growth of L. lactis and E. faecium isolates in the presence of simulated gastrointestinal fluid is elucidated. Ten milliliters of each isolate culture was collected by centrifugation at 8000×g at 4 °C for 10 min. Cell pellets were then washed twice with phosphate-buffered saline (PBS) at pH 7.4, followed by re-suspension in 50 ml of simulated gastric solutions (125 mM NaCl, 7 mM KCl, 45 mM NaHCO3, and 3 g/l pepsin; pH 2 and pH 3). To elucidate the effect of a food matrix on the isolates during gastric transit, the survival of both isolates in simulated gastric solution at pH 2 in the presence of skimmed milk (11% w/v) (Himedia, Mumbai, India) was also observed. All bacterial suspensions were then incubated at 37 °C with agitation. Samples were taken after 0, 90, and 180 min of incubation. After 180 min of incubation with simulated gastric solution, bacterial cells were harvested by centrifugation. Cell pellets were washed twice with PBS and re-suspended in simulated intestinal solution (0.1% w/v pancreatin (TCI, Chuo-ku, Tokyo, Japan) and 0.15% w/v oxgall bile salts (Neogen, Lansing, MI, USA); pH 8.0). The bacterial suspensions were continually incubated as described above. Samples of the bacterial suspensions were taken at 0, 90, and 180 min after incubation with simulated intestinal solution. Tenfold serial dilutions in PBS of each bacterial suspension were applied, and appropriated dilutions were spread on MRS agar plates. Cell viability was enumerated by counting the number of colony-forming units (CFU/ml) following overnight incubation at 37 °C [11].
Adhesion Assay
Human colon adenocarcinoma cell line Caco-2 cells (ATCC®HTB-37™) were cultured in Hyclone™ Dulbecco’s modified Eagle’s medium (DMEM) (GE Healthcare-Bioscience, Pittsburgh, PA, USA) supplemented with 10% Gibco™ Fetal Bovine Serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA) and 1% of Hyclone™ Penicillin-Streptomycin 100× solution (GE Healthcare-Bioscience, Pittsburgh, PA, USA) at 37 °C with 5% CO2 and humidity.
Caco-2 cells were seeded in a 24-well plate and cultured for 14 days until cells differentiated. Five hundred microliters of appropriate bacterial dilution (105 CFU/ml) were added to each well with a ratio of 10:1 (bacterial cells/Caco-2 cells) and incubated at 37 °C with 5% CO2 for 2 h. Cells were then washed with PBS thrice. Following the final wash, 500 μl of 0.1% Triton x-100 was added and pipetted vigorously to re-suspended Caco-2 cells with adhered bacterial cells. Tenfold serial dilutions of each mixture were done, and the appropriated dilutions were spread on MRS agar. Adhering bacteria were enumerated by counting CFU per milliliter following incubation at 37 °C for 24 h [12].
Cytotoxicity Assay
Caco-2 cells were seeded in a 96-well plate (3 × 104 cells/well) and incubated at 37 °C with 5% CO2 for 24 h. Medium was removed and 50 μl of DMEM was added to each well prior to the experiments. Fifty microliters of NCFS of E. faecium and L. lactis isolates and the CFS of CD630 with various total protein concentrations (100, 80, 40, 20, 10 μg/ml) were added into each well. The mixtures were incubated at 37 °C for 24 h. The mixtures were then removed, and 100 μl of 0.5 mg/ml of 3-(4,5-dimethylthiasol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (Merck, Kenilworth, NJ, USA) was added to each well, and the plate was further incubated for 2 h in darkness. The MTT solution was gently removed, 100 μl dimethyl sulfoxide (DMSO) (Merck, Kenilworth, NJ, USA) was added to solubilize formazan crystals, and the absorbance was measured at λ = 570 nm. DMEMs mixed with MRS or BHI were used as controls. Effects of LAB isolates against CD630 were also investigated under three different conditions: (1) coincubation: Caco-2 cells were incubated with 50 μl of 100 μg/ml NCFS of either E. faecium or L. lactis and 50 μl of 100 μg/ml CFS of CD630 following by incubation for 24 h; (2) protection: Caco-2 cells were pre-incubated with 50 μl of 100 μg/ml NCFS of either E. faecium or L. lactis for 2 h, followed by the addition of 50 μl of 100 μg/ml CFS of CD630, and the culture was further incubated for 24 h; and (3) prevention: Caco-2 cells were pre-incubated with 50 μl of 100 μg/ml CFS of CD630 for 2 h, followed by the addition of 50 μl of 100 μg/ml E. faecium or L. lactis, and further incubated for 24 h. DMEMs mixed with BHI and MRS were used as controls [13]. Experiments were conducted in triplicate. The Caco-2 cell viability was calculated using the following equation: % Cell viability = [(ODsample × 100)/ODcontrol].
Immunofluorescent Imaging of Tight Junctions
Caco-2 cells were cultured in a 24-well plate (1 × 105 cells/well) and incubated for 24 h. Cells were washed twice with PBS, and 500 μl of DMEM was added to each well followed by 500 μl of the NCFS of E. faecium, L. lactis, and the CFS of CD630 to a final concentration of 50 μg/ml total protein. Cells were incubated at 37 °C for 16 h, fixed with 4% v/v formaldehyde, permeabilized with 0.1% v/v Triton x-100, and washed thrice with PBS. Normal goat serum (5% v/v) (Merck, Kenilworth, NJ, USA) was used as a blocking solution. Primary antibodies against occludin (Merck, Kenilworth, NJ, USA) with the ratio of 1:100 were added, and cells were further incubated at 4 °C overnight. Cells were washed thrice with PBS, and FIT-C-conjugated secondary antibodies (1:100) (Merck, Kenilworth, NJ, USA) were added; cells were incubated at room temperature in darkness for 1 h. Cells were washed with PBS, followed by observation under a fluorescence microscope [14].
RT-qPCR Analysis
Caco-2 cells were seeded in a 6-well plate (106 cells/well) and cultured for 48 h. Five hundred microliters of DMEM was added to cells, followed by the addition of 500 μl of 100 μg/ml NCFS of E. faecium, L. lactis, or the CFS of CD630 into each well, and the plate was incubated for 16 h. The coincubation conditions, in which 500 μl of DMEM, 500 μl of 100 μg/ml NCFS of E. faecium or L. lactis, and 500 μl of 100 μg/ml CFS of CD630, were added to the same well, and were also prepared and incubated for 16 h. RNA extraction was performed using the illustra™ RNAspin Mini (GE Healthcare UK, Little Chalfont, Buckinghamshire, UK). Extracted RNA from all tested samples was quantified using NanoDrop Spectrophotometer at λ = 260 nm. To determine the purity of extracted RNA, the ratio of absorbance at λ = 260 and at λ = 280 nm (A260/A280) and the ratio of A260/A230 were also calculated. One microgram (1 μg) of each RNA sample was converted to cDNA using iScript™ Reverse Transcription Supermix for RT-qPCR (Bio-Rad, Hercules, CA, USA) following the manufacturer’s protocol and stored at − 20 °C until use. PCR was performed using 2× Taq Master Mix (Vivantis, Subang Jaya, Salangor Darul Ehsan, Malaysia) following the manufacturer’s protocol to obtain optimum conditions (Table 1). The expression level of IL-8 and TNF-α was investigated using Luna® Universal One-Step RT-qPCR Kit (NEB, Ipswich, MA, USA), while the expressions of β-actin were used as a control. The ΔΔCt method was used for determination of relative gene expression quantitation.
Statistical Analysis
All experiments were performed in triplicate, and the results were reported as the mean ± SEM of triplicated experiments. Data were analyzed using the Student’s t test with two-tailed distribution and considered statistically significant at a p value ≤ 0.05.
Results
Survival of LAB in Simulated Gastrointestinal Solution
Acid and bile salt tolerance are crucial properties for probiotic microorganisms which help them to survive through gastrointestinal tract. The survival of E. faecium and L. lactis in simulated gastrointestinal conditions was studied whether they can survive under acidic and alkaline condition. The result showed that the cell viability of E. faecium and L. lactis was drastically reduced during 90 min of incubation in simulated gastric solution at pH 2 (Fig. 1a, b). Both isolates survived in simulated gastric solution at pH 3 followed by intestinal solution at pH 8. However, L. lactis was more sensitive to these conditions, as its cell viability rapidly decreased compared to that of E. faecium. In addition, in the presence of skimmed milk added as a food matrix, both isolates subsequently demonstrated higher bacterial cell viability following incubation in simulated gastrointestinal solution.
Cell viability (log CFU/ml) of E. faecium (a) and L. lactis (b) incubated in simulated gastric solution at pH 2 (square) and pH 3 (circle), followed by simulated intestinal solution at pH 8; and gastric solution at pH 2 mixed with skimmed milk (triangle), followed by intestinal solution at pH 8. Bars represent standard deviation values
Adhesion Assay
The ability to adhere to intestinal epithelial cells is one of the most important properties for any probiotic bacteria in order to colonize in the gut mucosa. Caco-2 cells were used as a model of human colonic cells to study the adhesion properties of selected LAB. The result indicated that more than 60% of added E. faecium (61%) and L. lactis (67%) were able to adhere to Caco-2 cells (Fig. 2). There was no significant difference between the adhesive abilities of the isolates.
Cytotoxicity Assay
Evaluation of bacterial toxicity is necessary when those bacterial strains are considered to be ingested as probiotics. Any probiotic strains should not be harmful to the cells. Therefore, the NCFS from E. faecium and L. lactis and the CFS of CD630 were tested for their toxicity toward Caco-2 cells using an MTT assay. The results demonstrated that the NCFS of E. faecium and L. lactis were not toxic to Caco-2 cells, while the CFS of CD630 exhibited significant toxicity at increasing total protein concentrations (data not shown). The cytotoxicity of the NCFS of each LAB coincubated with the CFS of CD630 in different incubation patterns was also studied. The results demonstrated that the NCFS of L. lactis significantly improved Caco-2 cell viability compared to that of the NCFS of E. faecium when incubated with the CFS of CD630 (Fig. 3). Changes in the incubation patterns (coincubation, protection, and prevention) did not significantly affect Caco-2 cytotoxicity when mixed with NCFS of LAB isolates with CFS of CD630.
Cell viability of Caco-2 cells incubated with the NCFS of E. faecium or L. lactis combined with the CFS of CD630 for 24 h (co-incubation: E. faecium+CD630, L. lactis+CD630). The Caco-2 cells were pre-incubated with the NCFS of E. faecium or L. lactis for 2 h, followed by the addition of CFS of CD630 for 24 h (protection: E. faecium → CD630, L. lactis → CD630) and vice versa (prevention: CD630 → E. faecium, CD630 → L. lactis). Results are expressed as means ± SEM of triplicate experiments. The statistically significant difference from the control with the p value ≤ 0.05 and ≤ 0.01 was indicated by one asterisk (*) and double asterisks (**), respectively
Immunofluorescent Imaging of Tight Junction in Caco-2 Cell
Tight junction is a crucial component of an epithelial barrier. Microorganisms have to disrupt proteins in tight junction in order to penetrate the epithelial cells. Hence, the effects of the NCFS of E. faecium and L. lactis and the CFS of CD630 on tight junctions were observed by immunofluorescence staining of occludin, a transmembrane protein located at the tight junctions. The results demonstrated that the presence of MRS broth as a control treatment showed a morphological effect on Caco-2 cells (Fig. 4a). The NCFS of E. faecium and L. lactis showed minimal effects on tight junctions, as the structures of most cell membranes were still intact (Fig. 4b, c). Not surprisingly, compared to that of BHI broth as a control treatment (Fig. 4d), the CFS of CD630 affected the integrity of tight junctions by disrupting the structure of the cell membrane (Fig. 4e).
Effect of LAB and CD630 on Immunomodulation
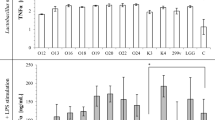
The level of pro-inflammatory cytokine expression in the Caco-2 cells, such as IL-8 and TNF-α, was investigated using RT-qPCR. Optimum conditions for β-actin, IL-8, and TNF-α expressions were achieved via PCR (data not shown). The results indicated that IL-8 and TNF-α were expressed at a low level when Caco-2 cells were incubated with the NCFS of E. faecium and L. lactis, while the CFS of CD630 significantly increased the level of IL-8 and TNF-α expressions in Caco-2 cells (Fig. 5a, b). The effects of the NCFS of either E. faecium or L. lactis coincubated with the CFS of CD630 were also observed. The presence of the NCFS of E. faecium and L. lactis significantly reduced the expression levels of IL-8 and TNF-α caused by the CFS of CD630.
Fold differences in pro-inflammatory cytokine expression; IL-8 (a) and TNF-α (b) after incubation with the NCFS of E. faecium, L. lactis, and the CFS of CD630. Results are expressed as means ± standard deviation for duplicate experiments. The statistically significant difference from the control with the p value ≤ 0.05 and ≤ 0.01 was indicated by one asterisk (*) and double asterisks (**), respectively
Discussion
Our findings indicate that E. faecium and L. lactis isolated from Thai fermented sausages possess desirable probiotic properties. Both were resistant to simulated gastrointestinal solution, one of the most common probiotic properties. Our results are in agreement with those of previous studies, which reported that some LAB isolated from vaginal microflora, as well as E. mundtii 50H isolated from traditional dairy, were tolerant of low pH, bile salts, and simulated digestive conditions in vitro [15, 16]. L. lactis isolated from sea pineapple kimchi also showed high resistance to acidic and bile salt conditions [17].
Following LAB survival through the stomach and small intestine, those bacteria should be able to adhere to colon epithelial cells and initiate colonization. Most LAB have high adhesive abilities, and their adhesion to Caco-2 cells was recently reported [15,16,17]. In the present study, more than 60% of added E. faecium and L. lactis isolates were able to adhere to Caco-2 cells. Previous research suggests that various strains of Lactobacillus and Bifidobacterium possess high adhesive ability to human intestinal cell lines and that proteinaceous components in their spent culture broth also increase adhesion ability [18,19,20]. However, the adhesive property of LAB may vary from strain to strain, as each strain has different cell-surface molecules that play role in adhesion ability.
Any probiotic strains should be harmless to hosts. In the present study, we used the NCFS of LAB to study their cytotoxic effect on Caco-2 cells. We demonstrated that the NCFS of E. faecium and L. lactis isolates were not toxic to Caco-2 cells but rather increased Caco-2 cell viability. Cytotoxicity of the CFS of CD630 was evaluated as well, and the results demonstrated that the CFS of CD630 was highly toxic to Caco-2 cells. Furthermore, the NCFS of E. faecium and L. lactis facilitated cell viability and demonstrated cell protection abilities when coincubated with the CFS of CD630. Recent studies support our findings that LAB can help protect cells from clostridial toxins. In one such study by Eden et al., L. rhamnosus GG was cocultured with C. difficile at different concentrations, and the CFS of cocultured bacteria were incubated with Vero cells. They found that L. rhamnosus GG reduced the cytotoxic effects of clostridial toxins compared to pure C. difficile supernatant [7].
Most pathogens cause the disruption of epithelial barriers, including C. difficile. The toxin A (TcdA) influenced F-actin and enhanced tight junction permeability in the T84 cell line [21]. The T84 cell line was treated with C. difficile reference toxin A (TcdA-10463) and toxin B (TcdB-10463), and both toxins altered tight junction proteins, which subsequently increased paracellular permeability causing cell death [22]. There is considerable evidence that LAB enhance the integrity of epithelial cells and as such protect these cells from pathogenic bacterial invasion. A previous study by Anderson et al. demonstrated that 19 tight junction-related genes were altered in response to L. plantarum MB452, and the intensity of the immuno-stained ZO-1, ZO-2, and occludin appeared higher in Caco-2 cells treated with L. plantarum MB452 [23]. In another study, the probiotic mixture VSL#3 was used to treat a dextran sodium sulfate (DSS)-induced colitis mouse model. Results demonstrated that the VSL#3 protected the epithelial barrier by maintaining the tight junction protein expression in this murine model [24]. Using immunofluorescence staining, we achieved parallel results indicating that NCFS of E. faecium and L. lactis did not affect the integrity of tight junction proteins in Caco-2 cells. In contrast, the CFS of CD630 observably damaged tight junction proteins, as the disruption of cell membranes was observed under a fluorescence microscope.
Another critical property of probiotics is immunomodulation of the host cell. Various strains of LAB are capable of activating immune responses against pathogens. During infection, pro-inflammatory cytokines are released and subsequently trigger cell inflammation. Injection of TcdA and TcdB into human intestinal xenografts caused histopathological changes and acute mucosal inflammation, and both toxins elevated levels of IL-8 gene expression and protein production [25]. Recent findings have demonstrated that some LAB alleviate inflammation. In one study by Tian et al., E. faecium HDRsEf1 live cells and CFS were used to treat porcine epithelial cells from the jejunum, and their anti-inflammatory effects against Escherichia coli (ETEC) were observed. Both live cells and CFS decreased the level of IL-8 gene expression [26]. Luerce et al. demonstrated that the supernatant of L. lactis NCDO2118 reduced the production of IL-8 and TNF-α in Caco-2 cells. Administration of these bacteria also ameliorated the histological damage in the colon of a DSS-induced colitis mouse model [27]. We obtained similar data that the NCFS of E. faecium and L. lactis reduced the level of IL-8 and TNF-α when coincubated with the CFS of CD630 in Caco-2 cells. As a result, E. faecium and L. lactis isolates exhibited anti-inflammatory effects against CD630.
In conclusion, E. faecium and L. lactis isolated from Thai fermented sausages are suitable candidates for use as probiotics, exhibiting such desirable qualities as high adhesion to and low cytotoxicity toward Caco-2 cells from the human intestinal epithelium, as well as cytoprotective and anti-inflammatory properties. These bacteria could be used as an alternative treatment to prevent CDI. However, additional study is required to determine the effect of using live E. faecium and L. lactis isolates in vivo.
References
Gareau MG, Sherman PM, Walker WA (2010) Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol 7(9):503–514. https://doi.org/10.1038/nrgastro.2010.117
Sherman PM, Ossa JC, Johnson-Henry K (2009) Unraveling mechanisms of action of probiotics. Nutr Clin Pract 24(1):10–14. https://doi.org/10.1177/0884533608329231
Deneve C, Janoir C, Poilane I, Fantinato C, Collignon A (2009) New trends in Clostridium difficile virulence and pathogenesis. Int J Antimicrob Agents 33(Suppl 1):S24–S28. https://doi.org/10.1016/s0924-8579(09)70012-3
Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP (2010) The role of toxin A and toxin B in Clostridium difficile infection. Nature 467(7316):711–713. https://doi.org/10.1038/nature09397
Voth DE, Ballard JD (2005) Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 18(2):247–263. https://doi.org/10.1128/cmr.18.2.247-263.2005
Kolling GL, Wu M, Warren CA, Durmaz E, Klaenhammer TR, Timko MP, Guerrant RL (2012) Lactic acid production by Streptococcus thermophilus alters Clostridium difficile infection and in vitro toxin a production. Gut Microbes 3(6):523–529. https://doi.org/10.4161/gmic.21757
Ephraim E, Schultz RD, Safdar N (2013) Lactobacillus rhamnosus GG protects cells from Clostridium difficile toxins. Br Microbiol Res J 3:165–175
Lee JS, Chung MJ, Seo JG (2013) In vitro evaluation of antimicrobial activity of lactic acid bacteria against Clostridium difficile. Toxicol Res 29(2):99–106. https://doi.org/10.5487/tr.2013.29.2.099
Suwanjinda D, Eames C, Panbangred W (2007) Screening of lactic acid bacteria for bacteriocins by microbiological and PCR methods. Biochem Mol Biol Educ 35(5):364–369. https://doi.org/10.1002/bmb.84
Nilsson WB, Paranjype RN, DePaola A, Strom MS (2003) Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J Clin Microbiol 41(1):442–446. https://doi.org/10.1128/JCM.41.1.442-446.2003
De Angelis M, Siragusa S, Berloco M, Caputo L, Settanni L, Alfonsi G, Amerio M, Grandi A, Ragni A, Gobbetti M (2006) Selection of potential probiotic lactobacilli from pig feces to be used as additives in pelleted feeding. Res Microbiol 157(8):792–801. https://doi.org/10.1016/j.resmic.2006.05.003
Todoriki K, Mukai T, Sato S, Toba T (2001) Inhibition of adhesion of food-borne pathogens to Caco-2 cells by Lactobacillus strains. J Appl Microbiol 91(1):154–159
Sevda ER, Koparal AT, Kivanc M (2015) Cytotoxic effects of various lactic acid bacteria on Caco-2 cells. Turk J Biol 39:23–30
Yu Q, Yuan L, Deng J, Yang Q (2015) Lactobacillus protects the integrity of intestinal epithelial barrier damaged by pathogenic bacteria. Front Cell Infect Microbiol 5:26. https://doi.org/10.3389/fcimb.2015.00026
Haghshenas B, Haghshenas M, Nami Y, Yari Khosroushahi A, Abdullah N, Barzegari A, Rosli R, Hejazi MS (2016) Probiotic assessment of Lactobacillus plantarum 15HN and Enterococcus mundtii 50H isolated from traditional dairies microbiota. Adv Pharm Bull 6(1):37–47. https://doi.org/10.15171/apb.2016.007
Nami Y, Abdullah N, Haghshenas B, Radiah D, Rosli R, Khosroushahi AY (2014) Probiotic assessment of Enterococcus durans 6HL and Lactococcus lactis 2HL isolated from vaginal microflora. J Med Microbiol 63(Pt 8):1044–1051. https://doi.org/10.1099/jmm.0.074161-0
Kawahara M, Nemoto M, Nakata T, Kondo S, Takahashi H, Kimura B, Kuda T (2015) Anti-inflammatory properties of fermented soy milk with Lactococcus lactis subsp. lactis S-SU2 in murine macrophage RAW264.7 cells and DSS-induced IBD model mice. Int Immunopharmacol 26(2):295–303. https://doi.org/10.1016/j.intimp.2015.04.004
Bernet MF, Brassart D, Neeser JR, Servin AL (1994) Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut 35(4):483–489
Chauviere G, Coconnier MH, Kerneis S, Fourniat J, Servin AL (1992) Adhesion of human Lactobacillus acidophilus strain LB to human enterocyte-like Caco-2 cells. J Gen Microbiol 138(Pt 8):1689–1696. https://doi.org/10.1099/00221287-138-8-1689
Gopal PK, Prasad J, Smart J, Gill HS (2001) In vitro adherence properties of Lactobacillus rhamnosus DR20 and Bifidobacterium lactis DR10 strains and their antagonistic activity against an enterotoxigenic Escherichia coli. Int J Food Microbiol 67(3):207–216
Hecht G, Pothoulakis C, LaMont JT, Madara JL (1988) Clostridium difficile toxin A perturbs cytoskeletal structure and tight junction permeability of cultured human intestinal epithelial monolayers. J Clin Invest 82(5):1516–1524. https://doi.org/10.1172/jci113760
Nusrat A, von Eichel-Streiber C, Turner JR, Verkade P, Madara JL, Parkos CA (2001) Clostridium difficile toxins disrupt epithelial barrier function by altering membrane microdomain localization of tight junction proteins. Infect Immun 69(3):1329–1336. https://doi.org/10.1128/iai.69.3.1329-1336.2001
Anderson RC, Cookson AL, McNabb WC, Park Z, McCann MJ, Kelly WJ, Roy NC (2010) Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation. BMC Microbiol 10:316. https://doi.org/10.1186/1471-2180-10-316
Mennigen R, Nolte K, Rijcken E, Utech M, Loeffler B, Senninger N, Bruewer M (2009) Probiotic mixture VSL#3 protects the epithelial barrier by maintaining tight junction protein expression and preventing apoptosis in a murine model of colitis. Am J Physiol Gastrointest Liver Physiol 296(5):G1140–G1149. https://doi.org/10.1152/ajpgi.90534.2008
Savidge TC, Pan WH, Newman P, O'Brien M, Anton PM, Pothoulakis C (2003) Clostridium difficile toxin B is an inflammatory enterotoxin in human intestine. Gastroenterology 125(2):413–420
Tian Z, Liu X (2016) Enterococcus faecium HDRsEf1 protects the intestinal epithelium and attenuates ETEC-induced IL-8 secretion in enterocytes. Mediat Inflamm 2016:7474306. https://doi.org/10.1155/2016/7474306
Luerce TD, Gomes-Santos AC, Rocha CS, Moreira TG, Cruz DN, Lemos L, Sousa AL, Pereira VB, de Azevedo M, Moraes K, Cara DC, LeBlanc JG, Azevedo V, Faria AMC, Miyoshi A (2014) Anti-inflammatory effects of Lactococcus lactis NCDO 2118 during the remission period of chemically induced colitis. Gut Pathog 6:33. https://doi.org/10.1186/1757-4749-6-33
Funding
This work was supported by the Faculty of Tropical Medicine Research Fund, Mahidol University, Thailand. Mrs. Panya Dowdell was supported by iBelieve2 scholarship from Wall Street English, Bangkok, Thailand.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Ethical Approval
This article does not contain any studies with human or animal subjects.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Dowdell, P., Chankhamhaengdecha, S., Panbangred, W. et al. Probiotic Activity of Enterococcus faecium and Lactococcus lactis Isolated from Thai Fermented Sausages and Their Protective Effect Against Clostridium difficile. Probiotics & Antimicro. Prot. 12, 641–648 (2020). https://doi.org/10.1007/s12602-019-09536-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-019-09536-7