Abstract
We propose a hypothesis that explains two apparently contradicting observations for the heterogeneity of the unfolded proteins. First, the line confocal method of the single-molecule Förster resonance energy transfer (sm-FRET) spectroscopy revealed that the unfolded proteins possess broad peaks in the FRET efficiency plot, implying the significant heterogeneity that lasts longer than milliseconds. Second, the fluorescence correlation method demonstrated that the unfolded proteins fluctuate in the time scale shorter than 100 ns. To formulate the hypothesis, we first summarize the recent consensus for the structure and dynamics of the unfolded proteins. We next discuss the conventional method of the sm-FRET spectroscopy and its limitations for the analysis of the unfolded proteins, followed by the advantages of the line confocal method that revealed the heterogeneity. Finally, we propose that the structural heterogeneity formed by the local clustering of hydrophobic residues modulates the distribution of the long-range distance between the labeled chromophores, resulting in the broadening of the peak in the FRET efficiency plot. A clustering of hydrophobic residues around the chromophore might further contribute to the broadening. The proposed clusters are important for the understanding of protein folding mechanism.

Similar content being viewed by others
References
Abaskharon RM, Gai F (2016) Meandering down the energy landscape of protein folding: are we there yet? Biophys J 110:1924–1932
Aznauryan M, Delgado L, Soranno A, Nettels D, Huang J, Labhardt AM, Grzesiek S, Schuler B (2016) Comprehensive structural and dynamical view of an unfolded protein from the combination of single-molecule FRET, NMR, and SAXS. Proc Natl Acad Sci U S A 113:E5389–E5398
Baxa MC, Haddadian EJ, Jumper JM, Freed KF, Sosnick TR (2014) Loss of conformational entropy in protein folding calculated using realistic ensembles and its implications for NMR-based calculations. Proc Natl Acad Sci U S A 111:15396–15401
Bernado P, Blanchard L, Timmins P, Marion D, Ruigrok RW, Blackledge M (2005) A structural model for unfolded proteins from residual dipolar couplings and small-angle x-ray scattering. Proc Natl Acad Sci U S A 102:17002–17007
Borgia A, Wensley BG, Soranno A, Nettels D, Borgia MB, Hoffmann A, Pfeil SH, Lipman EA, Clarke J, Schuler B (2012) Localizing internal friction along the reaction coordinate of protein folding by combining ensemble and single-molecule fluorescence spectroscopy. Nature Commun 3:1195 (9 pages)
Borgia A, Zheng W, Buholzer K, Borgia MB, Schüler A, Hofmann H, Soranno A, Nettels D, Gast K, Grishaev A, Best RB, Schuler B (2016) Consistent view of polypeptide chain expansion in chemical denaturants from multiple experimental methods. J Am Chem Soc 138:11714–11726
Bowler BE (2012) Residual structure in unfolded proteins. Curr Opin Struct Biol 22:4–13
Buckler DR, Haas E, Scheraga HA (1995) Analysis of the structure of ribonuclease A in native and partially denatured states by time-resolved noradiative dynamic excitation energy transfer between site-specific extrinsic probes. Biochemistry 34:15965–15978
Buscaglia M, Lapidus LJ, Eaton WA, Hofrichter J (2006) Effects of denaturants on the dynamics of loop formation in polypeptides. Biophys J 91:276–288
Chen H, Ahsan SS, Santiago-Berrios MB, Abruña HD, Webb WW (2010) Mechanisms of quenching of Alexa fluorophores by natural amino acids. J Am Chem Soc 132:7244–7245
Cho J-H, Meng W, Sato S, Kim EY, Schindelin H, Raleigh DP (2014) Energetically significant networks of coupled interactions within an unfolded protein. Proc Natl Acad Sci U S A 111:12079–12084
Dar TA, Schaeffer RD, Daggett V, Bowler BE (2011) Manifestations of native topology in the denatured state ensemble of Rhodopseudomonas palustris cytochrome c’. Biochemistry 50:1029–1041
Deniz AA, Dahan M, Grunwell JR, Ha T, Faulhaber AE, Chemla DS, Weiss S, Schultz PG (1999) Single-pair fluorescence resonance energy transfer on freely diffusing molecules: observation of Förster distance dependence and subpopulations. Proc Natl Acad Sci U S A 96:3670–3675
Deniz AA, Laurence TA, Beligere GS, Dahan M, Martin AB, Chemla DS, Dawson PE, Schultz PG, Weiss S (2000) Single-molecule protein folding: diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2. Proc Natl Acad Sci U S A 97:5179–5184
Dill KA, MacCallum JL (2012) The protein-folding problem, 50 years on. Science 338:1042–1046
Felitsky D, Lietzow MA, Dyson HJ, Wright PE (2008) Modeling transient collapsed states of an unfolded protein to provide insights into early folding events. Proc Natl Acad Sci U S A 105:6278–6283
Fitzkee NC, Rose GD (2004) Reassessing random-coil statistics in unfolded proteins. Proc Natl Acad Sci U S A 101:12497–12502
Fuertes G, Banterle N, Ruff KM, Chowdhury A, Mercadante D, Koehler C, Kachala M, Estrada Girona G, Milles S, Mishra A, Onck PR, Gräter F, Esteban-Martín S, Pappu RV, Svergun DI, Lemke EA (2017) Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs FRET measurements. Proc Natl Acad Sci USA 114:E6342–E6351
Hagen SJ, Hofrichter J, Szabo A, Eaton WA (1996) Diffusion-limited contact formation in unfolded cytochrome c: estimating the maximum rate of protein folding. Proc Natl Acad Sci U S A 93:11615–11617
Haran G (2012) How, when and why proteins collapse: the relation to folding. Curr Opin Struct Biol 22:14–20
Hoefling M, Lima N, Haenni D, Seidel CAM, Schuler B, Grubmüller H (2011) Structural heterogeneity and quantitative FRET efficiency distributions of polyprolines through a hybrid atomistic simulation and Monte Carlo approach. PLoS One 6:e19791 (19 pages)
Hoffmann H (2016) Understanding disordered and unfolded proteins using single-molecule FRET and polymer theory. Methods Appl Fluoresc 4:042003 (8 pages)
Hofmann H, Soranno A, Borgia A, Gast K, Nettels D, Schuler B (2012) Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc Natl Acad Sci U S A 109:16155–16160
Holden SJ, Uphoff S, Hohlbein J, Yadin D, Le Reste L, Britton OJ, Kapanidis AN (2010) Defining the limits of single-molecule FRET resolution in TIRF microscopy. Biophys J 99:3102–3111
Huang J-r, Grzesiek S (2010) Ensemble calculations of unstructured proteins constrained by RDC and PRE data: a case study of urea-denatured ubiquitin. J Am Chem Soc 132:694–705
Jensen MR, Zweckstetter M, Huang J-r, Blackledge M (2014) Exploring free-energy landscapes of intrinsically disordered proteins at atomic resolution using NMR spectroscopy. Chem Rev 114:6632–6660
Jha AK, Colubri A, Freed KF, Sosnick TR (2005) Statistical coil model of the unfolded state: resolving the reconciliation problem. Proc Natl Acad Sci U S A 102:13099–13104
Kalinin S, Sisamakis E, Magennis SW, Felekyan S, Seidel CAM (2010) On the origin of broadening of single-molecule FRET efficiency distributions beyond shot noise limits. J Phys Chem B 114:6197–6206
Klein-Seetharaman J, Oikawa M, Grimshaw SB, Wirmer J, Duchardt E, Ueda T, Imoto T, Smith LJ, Dobson CM, Schwalbe H (2002) Long-range interactions within a nonnative protein. Science 295:1719–1722
Kohn JE, Millett IS, Jacob J, Zagrovic B, Dillon TM, Cingel N, Dothager RS, Seifert S, Thiyagarajan P, Sosnick TR, Hasan MZ, Pande VS, Ruczinski I, Doniach S, Plaxco KW. (2004) Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. USA. 101, 12491–12496. Erratum in: (2005) Proc. Natl. Acad. Sci. USA. 102, 14475
Konuma T, Kimura T, Matsumoto S, Goto Y, Fujisawa T, Fersht AR, Takahashi S (2011) Time-resolved small-angle X-ray scattering study of the folding dynamics of barnase. J Mol Biol 405:1284–1294
Kuzmenkina EV, Heyes CD, Nienhaus GU (2005) Single-molecule Förster resonance energy transfer study of protein dynamics under denaturing conditions. Proc Natl Acad Sci U S A 102:15471–15476
Lapidus LJ, Eaton WA, Hofrichter J (2000) Measuring the rate of intramolecular contact formation in polypeptides. Proc Natl Acad Sci U S A 97:7220–7225
Maity H, Reddy G (2016) Folding of protein L with implications for collapse in the denatured state ensemble. J Am Chem Soc 138:2609–2616
Meier S, Grzesiek S, Blackledge M (2007) Direct observation of dipolar couplings and hydrogen bonds across a β-hairpin in 8 M urea. J Am Chem Soc 129:9799–9807
Meng W, Luan B, Lyle N, Pappu RV, Raleigh DP (2013) The denatured state ensemble contains significant local and long-range structure under native conditions: analysis of the N-terminal domain of ribosomal protein L9. Biochemistry 52:2662–2671
Merchant KA, Best RB, Louis JM, Gopich IV, Eaton WA (2007) Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations. Proc Natl Acad Sci U S A 104:1528–1533
Moglich A, Joder K, Kiefhaber T (2006) End-to-end distance distributions and intrachain diffusion constants in unfolded polypeptide chains indicate intramolecular hydrogen bond formation. Proc. Natl. Acad. Sci. USA. 103, 12394–12399. Erratum in: (2008) Proc. Natl. Acad. Sci. USA. 105, 6787
Nettels D, Gopich IV, Hoffmann A, Schuler B (2007) Ultrafast dynamics of protein collapse from single-molecule photon statistics. Proc Natl Acad Sci U S A 104:2655–2660
Ohnishi S, Lee AL, Edgell MH, Shortle D (2004) Direct demonstration of structural similarity between native and denatured eglin C. Biochemistry 43:4064–4070
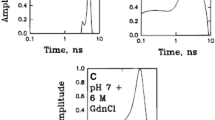
Oikawa H, Suzuki Y, Saito M, Kamagata K, Arai M, Takahashi S (2013) Microsecond dynamics of an unfolded protein by a line confocal tracking of single molecule fluorescence. Sci Rep 3:2151
Oikawa H, Kamagata K, Arai M, Takahashi S (2015) Complexity of the folding transition of the B domain of protein A revealed by the high-speed tracking of single-molecule fluorescence time series. J Phys Chem B 119:6081–6091
Oikawa H, Takahashi T, Kamonprasertsuk S, Takahashi S (2018) Microsecond resolved single-molecule FRET time series measurements based on line confocal optical system combined with hybrid photodetectors. Phys Chem Chem Phys 20:3277–3285
Plaxco KW, Millett IS, Segel DJ, Doniach S, Baker D (1999) Chain collapse can occur concomitantly with the rate-limiting step in protein folding. Nat Struct Biol 6:554–556
Riback JA, Bowman MA, Zmyslowski AM, Knoverek CR, Jumper JM, Hinshaw JR, Kaye EB, Freed KF, Clark PL, Sosnick TR (2017) Innovative scattering analysis shows that hydrophobic disordered proteins are expanded in water. Science 358:238–241
Saito M, Kamonprasertsuk S, Suzuki S, Nanatani K, Oikawa H, Kushiro K, Takai M, Chen PT, Chen EH, Chen RP, Takahashi S (2016) Significant heterogeneity and slow dynamics of the unfolded ubiquitin detected by the line confocal method of single-molecule fluorescence spectroscopy. J Phys Chem B 120:8818–8829
Sakurai K, Yagi M, Konuma T, Takahashi S, Nishimura C, Goto Y (2017) Non-native α-helices in the initial folding intermediate facilitate the ordered assembly of the β-barrel in β-lactoglobulin. Biochemistry 56:4799–4807
Schuler B, Lipman EA, Eaton WA (2002) Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature 419:743–747
Schuler B, Soranno A, Hofmann H, Nettels D (2016) Single-molecule FRET spectroscopy and the polymer physics of unfolded and intrinsically disordered proteins. Annu Rev Biophys 45:207–231
Sherman E, Haran G (2006) Coil-globule transition in the denatured state of a small protein. Proc Natl Acad Sci U S A 103:11539–11543
Shortle D (2002) The expanded denatured state: an ensemble of conformations trapped in a locally encoded topological space. Adv Protein Chem 62:1–23
Shortle D, Ackerman MS (2001) Persistence of native-like topology in a denatured protein in 8 M urea. Science 293:487–489
Song J, Gomes G-N, Shi T, Gradinaru CC, Chan HS (2017) Conformational heterogeneity and FRET data interpretation for dimensions of unfolded proteins. Biophys J 113:1012–1024
Soranno A, Longhi R, Bellini T, Buscaglia M (2009) Kinetics of contact formation and end-to-end distance distributions of swollen disordered peptides. Biophys J 96:1515–1528
Soranno A, Buchli B, Nettels D, Cheng RR, Müller-Späth S, Pfeil SH, Hoffmann A, Lipman EA, Makarov DE, Schuler B (2012) Quantifying internal friction in unfolded and intrinsically disordered proteins with single-molecule spectroscopy. Proc Natl Acad Sci U S A 109:17800–17806
Soranno A, Holla A, Dingfelder F, Nettels D, Makarov DE, Schuler B (2017) Integrated view of internal friction in unfolded proteins from single-molecule FRET, contact quenching, theory, and simulations. Proc Natl Acad Sci U S A 114:E1833–E1839
Sosnick TR, Barrick D (2011) The folding of single domain proteins: have we reached a consensus? Curr Opin Struct Biol 21:12–24
Sziegat F, Silvers R, Hähnke M, Jensen MR, Blackledge M, Wirmer-Bartoschek J, Schwalbe H (2012) Disentangling the coil: modulation of conformational and dynamic properties by site-directed mutation in the non-native state of hen egg white lysozyme. Biochemistry 51:3361–3372
Takahashi S, Kamagata K, Oikawa H (2016) Where the complex things are: single molecule and ensemble spectroscopic investigations of protein folding dynamics. Curr Opin Struct Biol 36:1–9
Tezuka-Kawakami T, Gell C, Brockwell DJ, Radford SE, Smith DA (2006) Urea-induced unfolding of the immunity protein Im9 monitored by spFRET. Biophys J 91:L42–L44
Udgaonkar JB (2013) Polypeptide chain collapse and protein folding. Arch Biochem Biophys 531:24–33
Waldauer SA, Bakajin O, Lapidus LJ (2010) Extremely slow intramolecular diffusion in unfolded protein L. Proc Natl Acad Sci U S A 107:13713–13717
Watkins HM, Simon AJ, Sosnick TR, Lipman EA, Hjelm RP, Plaxco KW (2015) Random coil negative control reproduces the discrepancy between scattering and FRET measurements of denatured protein dimensions. Proc Natl Acad Sci U S A 112:6631–6636
Xue Y, Skrynnikov NR (2011) Motion of a disordered polypeptide chain as studied by paramagnetic relaxation enhancements, 15N relaxation, and molecular dynamics simulations: how fast is segmental diffusion in denatured ubiquitin? J Am Chem Soc 133:14614–14628
Yoo TY, Meisburger SP, Hinshaw J, Pollack L, Haran G, Sosnick TR, Plaxco K (2012) Small-angle X-ray scattering and single-molecule FRET spectroscopy produce highly divergent views of the low-denaturant unfolded state. J Mol Biol 418:226–236
Acknowledgements
We dedicate this article to Prof. Fumio Arisaka. We thank our collaborators and the former and current members of our laboratory.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported by JSPS KAKENHI Grant Number JP25104007 (to S.T.) and JSPS KAKENHI Grant Number JP17K17608 (to H.O.).
Conflict of interest
Satoshi Takahashi declares that he has no conflict of interest. Aya Yoshida declares that she has no conflict of interest. Hiroyuki Oikawa declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
This article is part of a Special Issue on “Biomolecules to Bio-nanomachines-Fumio Arisaka 70th Birthday” edited by Damien Hall, Junichi Takagi and Haruki Nakamura
Rights and permissions
About this article
Cite this article
Takahashi, S., Yoshida, A. & Oikawa, H. Hypothesis: structural heterogeneity of the unfolded proteins originating from the coupling of the local clusters and the long-range distance distribution. Biophys Rev 10, 363–373 (2018). https://doi.org/10.1007/s12551-018-0405-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-018-0405-8