Abstract
Introduction
The combination of daclatasvir (DCV, pan-genotypic NS5A inhibitor) plus asunaprevir (ASV; NS3 protease inhibitor) is approved in Japan, Korea and other countries for the treatment of chronic hepatitis C virus (HCV) genotype (GT)-1. A high (~90 to 100%) sustained virologic response (SVR) with DCV/ASV therapy has been achieved by excluding patients infected with HCV GT-1b with baseline NS5A resistance-associated variants (RAVs) at L31 or Y93H detected by direct sequencing (DS). We set out to determine whether patients with minor variants at NS5A-L31 or -Y93H, detected by next-generation sequencing (NGS), impacted SVR rates with DCV/ASV therapy.
Methods
Baseline samples from 222 interferon (IFN)-ineligible/intolerant (N = 135) and prior non-responder (N = 87) patients infected with GT-1b who were treated with DCV/ASV for 24 weeks in the Phase 3 clinical study AI447026 were prepared for NGS (Ion-Torrent platform). The prevalence of baseline NS5A RAVs and their impact on SVR when observed at ≥1% by NGS in a patient’s virus population were examined. NGS and DS (sensitivity ≥20%) data were compared.
Results
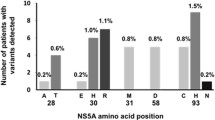
The prevalence of baseline NS5A RAVs at L31 or Y93H was 29% (63/219) and 18% (39/214) by NGS and DS, respectively. SVR24 rates were comparable in patients without observed baseline L31 or Y93H polymorphisms whether assessed by NGS (96%; 148/154) or by the less sensitive DS platform (95%; 164/173).
Conclusion
Optimal SVR rates (≥95%) to DCV/ASV treatment were achieved using DS to exclude patients infected with GT-1b with NS5A RAVs at L31 or Y93H representing ≥20% of their virus population. Exclusion by NGS of patients with minor variants in NS5A (<20%) did not enhance SVR rates. These results suggest that the presence of minor variants in NS5A does not appear to impact the overall SVR rate in patients with GT-1b treated with DCV/ASV.
Funding
This study was sponsored by Bristol-Myers Squibb.
Trial registration
ClinicalTrials.gov identifier: NCT01497834.



Similar content being viewed by others
References
Messina JP, Humphreys I, Flaxman A, et al. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61(1):77–87.
Seong MH, Kil H, Kim YS, et al. Clinical and epidemiological features of hepatitis C virus infection in South Korea: a prospective, multicenter cohort study. J Med Virol. 2013;85(10):1724–33.
Cho EJ, Jeong SH, Han BH, Lee SU, Yun BC, Park ET. Hepatitis C virus (HCV) genotypes and the influence of HCV subtype 1b on the progression of chronic hepatitis C in Korea: a single center experience. Clin Mol Hepatol. 2012;18(2):219–24.
Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–128.
Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol. 2014;61(1 Suppl):S45–57.
Gao M, Nettles RE, Belema M, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465(7294):96–100.
McPhee F, Sheaffer AK, Friborg J, et al. preclinical profile and characterization of the hepatitis C virus NS3 protease inhibitor asunaprevir (BMS-650032). Antimicrob Agents Chemother. 2012;56(10):5387–96.
Lok AS, Gardiner DF, Hézode C, et al. Randomized trial of daclatasvir and asunaprevir with or without PegIFN/RBV for hepatitis C virus genotype 1 null responders. J Hepatol. 2014;60(3):490–9.
Suzuki Y, Ikeda K, Suzuki F, et al. Dual oral therapy with daclatasvir and asunaprevir for patients with HCV genotype 1b infection and limited treatment options. J Hepatol. 2013;58(4):655–62.
Kumada H, Suzuki Y, Ikeda K, et al. Daclatasvir plus asunaprevir for chronic HCV genotype 1b infection. Hepatology. 2014;59(6):2083–91.
Manns M, Pol S, Jacobson IM, et al. All-oral daclatasvir plus asunaprevir for hepatitis C virus genotype 1b: a multinational, phase 3, multicohort study. Lancet. 2014;384(9954):1597–605.
Kumada H, Suzuki F, Suzuki Y, et al. Randomized comparison of daclatasvir + asunaprevir versus telaprevir + peginterferon/ribavirin in Japanese hepatitis C virus patients. J Gastroenterol Hepatol. 2016;31(1):14–22.
Wei L, Zhang M, Xu M, et al. A phase 3, open-label study of daclatasvir plus asunaprevir in Asian patients with chronic hepatitis C virus genotype 1b infection who are ineligible for or intolerant to interferon alfa therapies with or without ribavirin. J Gastroenterol Hepatol. 2016. doi:10.1111/jgh.13379 [Epub ahead of print].
Karino Y, Toyota J, Ikeda K, et al. Characterization of virologic escape in hepatitis C virus genotype 1b patients treated with the direct-acting antivirals daclatasvir and asunaprevir. J Hepatol. 2013;58(4):646–54.
McPhee F, Suzuki Y, Toyota J, et al. High sustained virologic response to daclatasvir plus asunaprevir in elderly and cirrhotic patients with hepatitis C virus genotype 1b without baseline NS5A polymorphisms. Adv Ther. 2015;32(7):637–49.
Uchida Y, Kouyama J, Naiki K, et al. Significance of variants associated with resistance to NS5A inhibitors in Japanese patients with genotype 1b hepatitis C virus infection as evaluated using cycling-probe real-time PCR combined with direct sequencing. J Gastroenterol. 2016;51(3):260–70.
Yoshimi S, Ochi H, Murakami E, et al. Rapid, sensitive, and accurate evaluation of drug resistant mutant (NS5A-Y93H) strain frequency in genotype 1b HCV by invader assay. PLoS One. 2015;10(6):e0130022.
Pallier C, Castéra L, Soulier A, et al. Dynamics of hepatitis B virus resistance to lamivudine. J Virol. 2006;80(2):643–53.
Palmer S, Kearney M, Maldarelli F, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43(1):406–13.
Hirotsu Y, Kanda T, Matsumura H, et al. HCV NS5A resistance-associated variants in a group of real-world Japanese patients chronically infected with HCV genotype 1b. Hepatol Int. 2015;9(3):424–30.
Zagordi O, Klein R, Däumer M, Beerenwinkel N. Error correction of next-generation sequencing data and reliable estimation of HIV quasispecies. Nucleic Acids Res. 2010;38(21):7400–9.
Beerenwinkel N, Zagordi O. Ultra-deep sequencing for the analysis of viral populations. Curr Opin Virol. 2011;1(5):413–8.
Murakami E, Imamura M, Hayes CN, et al. Ultradeep sequencing study of chronic hepatitis C virus genotype 1 infection in patients treated with daclatasvir, peginterferon, and ribavirin. Antimicrob Agents Chemother. 2014;58(4):2105–12.
Fridell RA, Qiu D, Wang C, Valera L, Gao M. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob Agents Chemother. 2010;54(9):3641–50.
Suzuki F, Sezaki H, Akuta N, et al. Prevalence of hepatitis C virus variants resistant to NS3 protease inhibitors or the NS5A inhibitor (BMS-790052) in hepatitis patients with genotype 1b. J Clin Virol. 2012;54(4):352–4.
Miura M, Maekawa S, Sato M, et al. Deep sequencing analysis of variants resistant to the non-structural 5A inhibitor daclatasvir in patients with genotype 1b hepatitis C virus infection. Hepatol Res. 2014;44(14):E360–7.
Kosaka K, Imamura M, Hayes CN, et al. Emergence of resistant variants detected by ultra-deep sequencing after asunaprevir and daclatasvir combination therapy in patients infected with hepatitis C virus genotype 1. J Viral Hepat. 2015;22(2):158–65.
Kinugasa H, Ikeda F, Takaguchi K, et al. Low frequency of drug-resistant virus did not affect the therapeutic efficacy in daclatasvir plus asunaprevir therapy in patients with chronic HCV genotype-1 infection. Antivir Ther. 2016;21(1):37–44.
Acknowledgments
All studies described and the analyses presented were sponsored by Bristol-Myers Squibb. The article processing charges and open access fee for this publication were funded by Bristol-Myers Squibb. All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this manuscript, take responsibility for the integrity of the work as a whole, and have given final approval to the version to be published.
Disclosures
Dennis Hernandez, Fei Yu, Xin Huang, Stefan Kirov, Saumya Pant, and Fiona McPhee are employees and stock holders of Bristol-Myers Squibb.
Compliance with Ethics Guidelines
The AI447026 study was approved by the Institutional Review Board at each participating site and was conducted in compliance with the Declaration of Helsinki, Good Clinical Practice guidelines, and local regulatory requirements. All patients provided written informed consent.
Author information
Authors and Affiliations
Corresponding author
Additional information
Enhanced content
To view enhanced content for this article go to http://www.medengine.com/Redeem/46D4F0601E311BFC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hernandez, D., Yu, F., Huang, X. et al. Impact of Pre-existing NS5A-L31 or -Y93H Minor Variants on Response Rates in Patients Infected with HCV Genotype-1b Treated with Daclatasvir/Asunaprevir. Adv Ther 33, 1169–1179 (2016). https://doi.org/10.1007/s12325-016-0354-1
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12325-016-0354-1