Abstract
Background
Majority of the BRCA1 and BRCA2 mutations are associated with the risk of sporadic and familial breast cancer. Since these genes are significant in DNA repair mechanisms, we focused homology-directed DNA repair (HDDR) and BRCA complex.
Methods
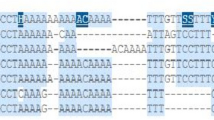
We selected BRCA1 variant (rs80356932, 4491C/T) and BRCA2 variant (rs80359182, 319T/C) from the interaction region of BRCA complex and studied in 100 breast cancer patients and 100 controls using tetra-ARMS-PCR.
Results
Here we show that BRCA1 and BRCA2 variants are significantly associated with high breast cancer risk (BRCA1 rs80356932; Genotype T/T OR 8.66, 95% CI 3.16–23.71, p < 0.0001; Allele-T, OR 2.48, 95% CI 1.62–3.81, p < 0.0001 and BRCA2 rs80359182; Genotype C/C OR 4.32, 95% CI 1.95–9.53, p = 0.0001; Allele-C, OR 2.19, 95% CI 1.43–3.34, p = 0.0002). Additionally, bioinformatics analysis showed that BRCA2-tryptophan > arginine substitutions result in altered interaction of BRCA1/PALB2/BRCA2/protein complex and impaired HDDR pathway. We also observed that breast cancer risk was significantly increased in over-weighted and obese women.
Conclusions
Our results indicate that high risk of breast cancer is significantly associated with BRCA1 and BRCA2 variants, and mutations may alter the protein interactions of BRCA complex that results in tumor genesis.

Similar content being viewed by others
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA. 2015;65(2):87–108.
Khan RT, Siddique A, Shahid N, Khokher S, Fatima W. Breast cancer risk associated with genes encoding DNA repair MRN complex: a study from Punjab, Pakistan. Breast Cancer. 2018;25(3):350–5. https://doi.org/10.1007/s12282-018-0837-9.
Bhurgri Y. Karachi cancer registry data—implications for the national cancer control program of Pakistan. Asian Pac J Cancer Prev. 2004;5(1):77–82.
Shaukat U, Ismail M, Mehmood N. Epidemiology, major risk factors and genetic predisposition for breast cancer in the Pakistani population. Asian Pac J Cancer Prev. 2013;14(10):5625–9.
Rosen M, Pishvaian EJ M. Targeting the BRCA1/2 tumor suppressors. Curr Drug Targets. 2014;15(1):17–31.
Aparicio T, Baer R, Gautier J. DNA double-strand break repair pathway choice and cancer. DNA Repair. 2014;19:169–75.
Kleibl Z, Kristensen VN. Women at high risk of breast cancer: molecular characteristics, clinical presentation and management. Breast. 2016;28:136–44.
Prodosmo A, Buffone A, Mattioni M, Barnabei A, Persichetti A, De Leo A, Appetecchia M, Nicolussi A, Coppa A, Sciacchitano S. Detection of ATM germline variants by the p53 mitotic centrosomal localization test in BRCA1/2-negative patients with early-onset breast cancer. J Exp Clin Cancer Res. 2016;35(1):135.
Kapoor NS, Curcio LD, Blakemore CA, Bremner AK, McFarland RE, West JG, Banks KC. Multigene panel testing detects equal rates of pathogenic BRCA1/2 mutations and has a higher diagnostic yield compared to limited BRCA1/2 analysis alone in patients at risk for hereditary breast cancer. Ann Surg Oncol. 2015;22(10):3282–8.
Norquist BM, Harrell MI, Brady MF, Walsh T, Lee MK, Gulsuner S, Bernards SS, Casadei S, Yi Q, Burger RA. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2(4):482–90.
Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7(4):a016600.
Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11(2):138.
Eroles P. Members of the BRCA1 complexes as new susceptibility genes for breast cancer. Transl Cancer Res. 2013;2(2):103–5.
Daley JM, Kwon Y, Niu H, Sung P. Focus: 50 years of DNA repair: the Yale symposium reports: investigations of homologous recombination pathways and their regulation. Yale J Biol Med. 2013;86(4):453.
Shahid T, Soroka J, Kong EH, Malivert L, McIlwraith MJ, Pape T, West SC, Zhang X. Structure and mechanism of action of the BRCA2 breast cancer tumor suppressor. Nat Struct Mol Biol. 2014;21(11):962.
Holloman WK. Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol. 2011;18(7):748.
Liu J, Doty T, Gibson B, Heyer W-D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17(10):1260.
Al Abo M, Dejsuphong D, Hirota K, Yonetani Y, Yamazoe M, Kurumizaka H, Takeda S. Compensatory functions and Interdependency of the DNA-binding domain of BRCA2 with the BRCA1–PALB2–BRCA2 complex. Cancer Res. 2014;74(3):797–807.
Oliver AW, Swift S, Lord CJ, Ashworth A, Pearl LH. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009;10(9):990–6.
Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19(6):524–9.
Park J-Y, Zhang F, Andreassen PR. ALB2: the hub of a network of tumor suppressors involved in DNA damage responses. Biochim Biophys Acta. 2014;1:263–75.
Apostolou P, Fostira F. Hereditary breast cancer: the era of new susceptibility genes. Biomed Res Int. 2013;2013:747318.
Ding YC, Steele L, Kuan C-J, Greilac S, Neuhausen SL. Mutations in BRCA2 and PALB2 in male breast cancer cases from the United States. Breast Cancer Res Treat. 2011;126(3):771–8.
Elia AE, Elledge SJ. BRCA1 as tumor suppressor: lord without its RING? Breast Cancer Res. 2012;14(2):306.
Kasparek TR. Humphrey TC DNA double-strand break repair pathways, chromosomal rearrangements and cancer. Semin Cell Dev Biol. 2011;8:886–97.
Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584(17):3703–8.
Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12(1):68.
Adank M, van Mil S, Gille J, Waisfisz Q, Meijers-Heijboer H. PALB2 analysis in BRCA2-like families. Breast Cancer Res Treat. 2011;127(2):357–62.
Lee EY, Muller WJ. Oncogenes and tumor suppressor genes. Cold Spring Harb Perspect Biol. 2010;2(10):a003236.
Anantha RW, Alcivar AL, Ma J, Cai H, Simhadri S, Ule J, König J, Xia B. Requirement of heterogeneous nuclear ribonucleoprotein C for BRCA gene expression and homologous recombination. PLoS One. 2013;8(4):e61368.
Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204.
Kang PCE, Phuah SY, Sivanandan K, Kang IN, Thirthagiri E, Liu JJ, Hassan N, Yoon S-Y, Thong MK, Hui M. Recurrent mutation testing of BRCA1 and BRCA2 in Asian breast cancer patients identify carriers in those with presumed low risk by family history. Breast Cancer Res Treat. 2014;144(3):635–42.
Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22(6):719–29.
Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001;29(17):e88–8.
Ikeda N, Miyoshi Y, Yoneda K, Shiba E, Sekihara Y, Kinoshita M, Noguchi S. Frequency of BRCA1 and BRCA2 germline mutations in Japanese breast cancer families. Int J Cancer. 2001;91(1):83–8.
Rasmussen SA, Overman J, Thomson SA, Colman SD, Abernathy CR, Trimpert RE, Moose R, Virdi G, Roux K, Bauer M. Chromosome 17 loss-of-heterozygosity studies in benign and malignant tumors in neurofibromatosis type 1. Genes Chromosom Cancer. 2000;28(4):425–31.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
About this article
Cite this article
Abbas, S., Siddique, A., Shahid, N. et al. Breast cancer risk associated with BRCA1/2 variants in the Pakistani population. Breast Cancer 26, 365–372 (2019). https://doi.org/10.1007/s12282-018-0932-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-018-0932-y