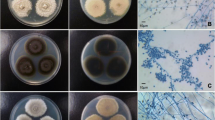
Abstract
Purpose of review
Black yeast-like fungi are capable of causing a wide range of infections, including invasive disease. The diagnosis of infections caused by these species can be problematic. We review the changes in the nomenclature and taxonomy of these fungi, and methods used for detection and species identification that aid in diagnosis.
Recent findings
Molecular assays, including DNA barcode analysis and rolling circle amplification, have improved our ability to correctly identify these species. A proteomic approach using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has also shown promising results. While progress has been made with molecular techniques using direct specimens, data are currently limited.
Summary
Molecular and proteomic assays have improved the identification of black yeast-like fungi. However, improved molecular and proteomic databases and better assays for the detection and identification in direct specimens are needed to improve the diagnosis of disease caused by black yeast-like fungi.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Nachman S, Alpan O, Malowitz R, Spitzer ED. Catheter-associated fungemia due to Wangiella (Exophiala) dermatitidis. J Clin Microbiol. 1996;34(4):1011–3.
Al-Obaid I, Ahmad S, Khan ZU, Dinesh B, Hejab HM. Catheter-associated fungemia due to Exophiala oligosperma in a leukemic child and review of fungemia cases caused by Exophiala species. Eur J Clin Microbiol Infect Dis. 2006;25(11):729–32.
Nucci M, Akiti T, Barreiros G, Silveira F, Revankar SG, Wickes BL, et al. Nosocomial outbreak of Exophiala jeanselmei fungemia associated with contamination of hospital water. Clin Infect Dis. 2002;34(11):1475–80.
• Seyedmousavi S, Netea MG, Mouton JW, Melchers WJ, Verweij PE, de Hoog GS. Black yeasts and their filamentous relatives: principles of pathogenesis and host defense. Clin Microbiol Rev. 2014;27(3):527–42. Comprehensive review of the black yeasts and their filamentous relatives.
Ventin M, Ramirez C, Garau J. Exophiala dermatitidis de Hoog from a valvular aortal prothesis. Mycopathologia. 1987;99(1):45–6.
Patel AK, Patel KK, Darji P, Singh R, Shivaprakash MR, Chakrabarti A. Exophiala dermatitidis endocarditis on native aortic valve in a postrenal transplant patient and review of literature on E. dermatitidis infections. Mycoses. 2013;56(3):365–72.
•• Revankar SG, Baddley JW, SCA C, Kauffman CA, Slavin M, Vazquez JA, et al. A Mycoses Study Group international prospective study of phaeohyphomycosis: an analysis of 99 proven/probable cases. Open Forum Infect Dis. 2017;4(4) https://doi.org/10.1093/ofid/ofx200. International case registry of patients with proven or probable phaeohyphomycosis that included several cases of infections caused by black yeast-like fungi and their filamentous relatives, including localized superficial and deep infections, and disseminated disease.
Gschwend A, Degot T, Denis J, Sabou AM, Jeung MY, Zapata E, et al. Brain abscesses caused by Cladophialophora bantiana in a lung transplant patient: a case report and review of the literature. Transpl Infect Dis. 2017;19(6)
Chakrabarti A, Kaur H, Rudramurthy SM, Appannanavar SB, Patel A, Mukherjee KK, et al. Brain abscess due to Cladophialophora bantiana: a review of 124 cases. Med Mycol. 2016;54(2):111–9.
Garzoni C, Markham L, Bijlenga P, Garbino J. Cladophialophora bantiana: a rare cause of fungal brain abscess. Clinical aspects and new therapeutic options. Med Mycol. 2008;46(5):481–6.
Doymaz MZ, Seyithanoglu MF, Hakyemez I, Gultepe BS, Cevik S, Aslan T. A case of cerebral phaeohyphomycosis caused by Fonsecaea monophora, a neurotropic dematiaceous fungus, and a review of the literature. Mycoses. 2015;58(3):187–92.
Koo S, Klompas M, Marty FM. Fonsecaea monophora cerebral phaeohyphomycosis: case report of successful surgical excision and voriconazole treatment and review. Med Mycol. 2010;48(5):769–74.
de Hoog GS, Vicente VA, Najafzadeh MJ, Harrak MJ, Badali H, Seyedmousavi S. Waterborne Exophiala species causing disease in cold-blooded animals. Persoonia. 2011;27:46–72.
Vicente VA, Orelis-Ribeiro R, Najafzadeh MJ, Sun J, Guerra RS, Miesch S, et al. Black yeast-like fungi associated with lethargic crab disease (LCD) in the mangrove-land crab, Ucides cordatus (Ocypodidae). Vet Microbiol. 2012;158(1–2):109–22.
Morrell M, Fraser VJ, Kollef MH. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob Agents Chemother. 2005;49(9):3640–5.
Garey KW, Rege M, Pai MP, Mingo DE, Suda KJ, Turpin RS, et al. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: a multi-institutional study. Clin Infect Dis. 2006;43(1):25–31.
Patterson TF, Thompson GR 3rd, Denning DW, Fishman JA, Hadley S, Herbrecht R, et al. Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis. 2016;63(4):e1-e60.
Rogers SO, McKemy JM, Wang CJK. Molecular assessment of Exophiala and related hyphomycetes. Stud Mycol. 1999;43:122–32.
•• Teixeira MM, Moreno LF, Stielow BJ, Muszewska A, Hainaut M, Gonzaga L, et al. Exploring the genomic diversity of black yeasts and relatives (Chaetothyriales, Ascomycota). Stud Mycol. 2017;86:1–28. Large, comprehensive study of the genomics of the black yeasts.
Gueidan C, Aptroot A, Caceres MED, Badali H, Stenroos S. A reappraisal of orders and families within the subclass Chaetothyriomycetidae (Eurotiomycetes, Ascomycota). Mycol Prog. 2014;13(4):1027–39.
Badali H, Bonifaz A, Barron-Tapia T, Vazquez-Gonzalez D, Estrada-Aguilar L, Oliveira NM, et al. Rhinocladiella aquaspersa, proven agent of verrucous skin infection and a novel type of chromoblastomycosis. Med Mycol. 2010;48(5):696–703.
de Hoog GS, Chaturvedi V, Denning DW, Dyer PS, Frisvad JC, Geiser D, et al. Name changes in medically important fungi and their implications for clinical practice. J Clin Microbiol. 2015;53(4):1056–62.
Honbo S, Padhye AA, Ajello L. The relationship of Cladosporium carrionii to Cladophialophora ajelloi. Sabouraudia. 1984;22(3):209–18.
De Hoog GS, Attili-Angelis D, Vicente VA, Van Den Ende AH, Queiroz-Telles F. Molecular ecology and pathogenic potential of Fonsecaea species. Med Mycol. 2004;42(5):405–16.
Badali H, Gueidan C, Najafzadeh MJ, Bonifaz A, van den Ende AH, de Hoog GS. Biodiversity of the genus Cladophialophora. Stud Mycol. 2008;61:175–91.
Gao L, Ma Y, Zhao W, Wei Z, Gleason ML, Chen H, et al. Three new species of Cyphellophora (Chaetothyriales) associated with sooty blotch and flyspeck. PLoS One. 2015;10(9):e0136857.
McGinnis MR. Wangiella dermatitidis, a correction. Stud Mycol. 1977;15:367–9.
Kidd S, Halliday C, Alexiou H, Ellis D. Descriptions of medical fungi. www.mycology.adelaide.edu.au: CutCut Digital; 2016.
de Hoog GS, Mayser P, Haase G, Horre R, Horrevorts AM. A new species, Phialophora europaea, causing superficial infections in humans. Mycoses. 2000;43(11–12):409–16.
Arzanlou M, Groenewald JZ, Gams W, Braun U, Shin HD, Crous PW. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud Mycol. 2007;58:57–93.
Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson RA, Seifert KA, et al. The Amsterdam declaration on fungal nomenclature. IMA Fungus. 2011;2(1):105–12.
McNeill J, Barrie FR, Buck WR, Demoulin V, Greuter W, Hawksworth DL, et al. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code). In: Regnum Vegetabile, vol. 154. Konigstein: Koeltz Sci Books; 2012.
de Hoog GS, Gueho E, Masclaux F, Gerrits van den Ende AH, Kwon-Chung KJ, McGinnis MR. Nutritional physiology and taxonomy of human-pathogenic Cladosporium-Xylohypha species. J Med Vet Mycol. 1995;33(5):339–47.
McGinnis MR. Wangiella, a new genus to accommodate Hormiscium dermatitidis. Mycotaxon. 1977;5:122–33.
Carrion AL. Preliminary report on a new clinical type of disease caused by Hormodendrum compactum, nov. sp. Puerto Rico J Publ Health Trop Med. 1935;10:543–5.
Attili DS, De Hoog GS, rDNA-RFLP P-KAA. ITS1 sequencing of species of the genus Fonsecaea, agents of chromoblastomycosis. Med Mycol. 1998;36(4):219–25.
Najafzadeh MJ, Sun J, Vicente V, Xi L, van den Ende AH, de Hoog GS. Fonsecaea nubica sp. nov, a new agent of human chromoblastomycosis revealed using molecular data. Med Mycol. 2010;48(6):800–6.
de Hoog GS. Rhinocladiella and allied genera. Stud Mycol. 1977;15:1–140.
Heinrichs G, de Hoog GS, Haase G. Barcode identifiers as a practical tool for reliable species assignment of medically important black yeast species. J Clin Microbiol. 2012;50(9):3023–30.
de Hoog GS, Takeo K, Yoshida S, Gottlich E, Nishimura K, Miyaji M. Pleoanamorphic life cycle of Exophiala (Wangiella) dermatitidis. Antonie Van Leeuwenhoek. 1994;65(2):143–53.
White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press, Inc.; 1990. p. 315–22.
Raja HA, Miller AN, Pearce CJ, Oberlies NH. Fungal identification using molecular tools: a primer for the natural products research community. J Nat Prod. 2017;80(3):756–70.
Lau A, Chen S, Sorrell T, Carter D, Malik R, Martin P, et al. Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J Clin Microbiol. 2007;45(2):380–5.
Zupancic J, Novak Babic M, Zalar P, Gunde-Cimerman N. The black yeast Exophiala dermatitidis and other selected opportunistic human fungal pathogens spread from dishwashers to kitchens. PLoS One. 2016;11(2):e0148166.
Gerritis van den Ende AH, de Hoog GS. Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana. Stud Mycol. 1999;43:151–62.
Najafzadeh MJ, Dolatabadi S, Saradeghi Keisari M, Naseri A, Feng P, de Hoog GS. Detection and identification of opportunistic Exophiala species using the rolling circle amplification of ribosomal internal transcribed spacers. J Microbiol Methods. 2013;94(3):338–42.
Sun J, Najafzadeh MJ, Zhang J, Vicente VA, Xi L, de Hoog GS. Molecular identification of Penicillium marneffei using rolling circle amplification. Mycoses. 2011;54(6):e751–9.
Nagano Y, Elborn JS, Millar BC, Goldsmith CE, Rendall J, Moore JE. Development of a novel PCR assay for the identification of the black yeast, Exophiala (Wangiella) dermatitidis from adult patients with cystic fibrosis (CF). J Cyst Fibros. 2008;7(6):576–80.
Libert X, Chasseur C, Packeu A, Bureau F, Roosens NH, De Keersmaecker SJ. A molecular approach for the rapid, selective and sensitive detection of Exophiala jeanselmei in environmental samples: development and performance assessment of a real-time PCR assay. Appl Microbiol Biotechnol. 2016;100(3):1377–92.
Saenz AJ, Petersen CE, Valentine NB, Gantt SL, Jarman KH, Kingsley MT, et al. Reproducibility of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for replicate bacterial culture analysis. Rapid Commun Mass Spectrom. 1999;13(15):1580–5.
Walker J, Fox AJ, Edwards-Jones V, Gordon DB. Intact cell mass spectrometry (ICMS) used to type methicillin-resistant Staphylococcus aureus: media effects and inter-laboratory reproducibility. J Microbiol Methods. 2002;48(2–3):117–26.
Bernardo K, Pakulat N, Macht M, Krut O, Seifert H, Fleer S, et al. Identification and discrimination of Staphylococcus aureus strains using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics. 2002;2(6):747–53.
Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49(4):543–51.
• Rychert J, Slechta ES, Barker AP, Miranda E, Babady NE, Tang YW, et al. Multicenter evaluation of the Vitek MS v3.0 System for the identification of filamentous fungi. J Clin Microbiol. 2018;56(2) https://doi.org/10.1128/JCM.01353-17. Multi-center study evaluating the clinical utility, including accuracy and reproducibility, of a MALDI-TOF MS diagnostic assay for the identification of medically important molds.
Nomura F. Proteome-based bacterial identification using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS): a revolutionary shift in clinical diagnostic microbiology. Biochim Biophys Acta. 2015;1854(6):528–37.
Croxatto A, Prod’hom G, Greub G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev. 2012;36(2):380–407.
Ryzhov V, Fenselau C. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal Chem. 2001;73(4):746–50.
Parisi D, Magliulo M, Nanni P, Casale M, Forina M, Roda A. Analysis and classification of bacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and a chemometric approach. Anal Bioanal Chem. 2008;391(6):2127–34.
Valentine N, Wunschel S, Wunschel D, Petersen C, Wahl K. Effect of culture conditions on microorganism identification by matrix-assisted laser desorption ionization mass spectrometry. Appl Environ Microbiol. 2005;71(1):58–64.
Fraser M, Borman AM, Johnson EM. Rapid and robust identification of the agents of black-grain mycetoma by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2017;55(8):2521–8.
Kondori N, Erhard M, Welinder-Olsson C, Groenewald M, Verkley G, Moore ER. Analyses of black fungi by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS): species-level identification of clinical isolates of Exophiala dermatitidis. FEMS Microbiol Lett. 2015;362(1):1–6.
Ozhak-Baysan B, Ogunc D, Dogen A, Ilkit M, de Hoog GS. MALDI-TOF MS-based identification of black yeasts of the genus Exophiala. Med Mycol. 2015;53(4):347–52.
Borman AM, Fraser M, Szekely A, Larcombe DE, Johnson EM. Rapid identification of clinically relevant members of the genus Exophiala by matrix-assisted laser desorption ionization-time of flight mass spectrometry and description of two novel species, Exophiala campbellii and Exophiala lavatrina. J Clin Microbiol. 2017;55(4):1162–76.
Odabasi Z, Paetznick VL, Rodriguez JR, Chen E, McGinnis MR, Ostrosky-Zeichner L. Differences in beta-glucan levels in culture supernatants of a variety of fungi. Med Mycol. 2006;44(3):267–72.
Odabasi Z, Mattiuzzi G, Estey E, Kantarjian H, Saeki F, Ridge RJ, et al. Beta-D-glucan as a diagnostic adjunct for invasive fungal infections: validation, cutoff development, and performance in patients with acute myelogenous leukemia and myelodysplastic syndrome. Clin Infect Dis. 2004;39(2):199–205.
•• Halliday CL, Kidd SE, Sorrell TC, Chen SC. Molecular diagnostic methods for invasive fungal disease: the horizon draws nearer? Pathology. 2015;47(3):257–69. Comprehensive review of molecular diagnostic assays for the detection and identificaiton of fungi, including cultures and direct specimens.
Gomez CA, Budvytiene I, Zemek AJ, Banaei N. Performance of targeted fungal sequencing for culture-independent diagnosis of invasive fungal disease. Clin Infect Dis. 2017;65(12):2035–41.
Hall L, Le Febre KM, Deml SM, Wohlfiel SL, Wengenack NL. Evaluation of the Yeast Traffic Light PNA FISH probes for identification of Candida species from positive blood cultures. J Clin Microbiol. 2012;50(4):1446–8.
Stone NR, Gorton RL, Barker K, Ramnarain P, Kibbler CC. Evaluation of PNA-FISH Yeast Traffic Light for rapid identification of yeast directly from positive blood cultures and assessment of clinical impact. J Clin Microbiol. 2013;51(4):1301–2.
Altun O, Almuhayawi M, Ullberg M, Ozenci V. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol. 2013;51(12):4130–6.
Mylonakis E, Clancy CJ, Ostrosky-Zeichner L, Garey KW, Alangaden GJ, Vazquez JA, et al. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin Infect Dis. 2015;60(6):892–9.
Neely LA, Audeh M, Phung NA, Min M, Suchocki A, Plourde D, et al. T2 magnetic resonance enables nanoparticle-mediated rapid detection of candidemia in whole blood. Sci Transl Med. 2013;5(182):182ra54.
Massire C, Buelow DR, Zhang SX, Lovari R, Matthews HE, Toleno DM, et al. PCR followed by electrospray ionization mass spectrometry for broad-range identification of fungal pathogens. J Clin Microbiol. 2013;51(3):959–66.
Simner PJ, Uhl JR, Hall L, Weber MM, Walchak RC, Buckwalter S, et al. Broad-range direct detection and identification of fungi by use of the PLEX-ID PCR-electrospray ionization mass spectrometry (ESI-MS) system. J Clin Microbiol. 2013;51(6):1699–706.
Shin JH, Ranken R, Sefers SE, Lovari R, Quinn CD, Meng S, et al. Detection, identification, and distribution of fungi in bronchoalveolar lavage specimens by use of multilocus PCR coupled with electrospray ionization/mass spectrometry. J Clin Microbiol. 2013;51(1):136–41.
Ozenci V, Patel R, Ullberg M, Stralin K. Demise of polymerase chain reaction/electrospray ionization-mass spectrometry as an infectious diseases diagnostic tool. Clin Infect Dis. 2018;66(3):452–5.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Connie F. Cañete-Gibas declares no conflict of interest.
Nathan P. Wiederhold has received grants from bioMerieux, Astellas, Pfizer, Merck, Viamet, F2G, MOE Medical Devices, and Cidara and has received travel reimbursement from Gilead.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Advances in Diagnosis of Invasive Fungal Infections
Rights and permissions
About this article
Cite this article
Cañete-Gibas, C.F., Wiederhold, N.P. The Black Yeasts: an Update on Species Identification and Diagnosis. Curr Fungal Infect Rep 12, 59–65 (2018). https://doi.org/10.1007/s12281-018-0314-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12281-018-0314-0