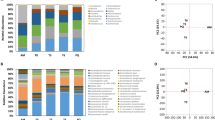
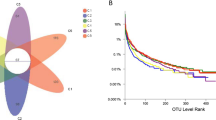
Abstract
White colony-forming yeasts (WCFYs) often appear in fermented foods, depending on the storage method. Despite the ongoing research on fermented foods, the community and genome features of WCFYs have not been well studied. In this study, the community structures of WCFYs on fermented vegetables (kimchi) prepared with various raw materials were investigated using deep sequencing. Only eight operational taxonomic units (OTUs) were detected, indicating that the community structure of WCFYs on kimchi is very simple. The five most abundant OTUs represented Pichia kluyveri, Yarrowia lipolytica, Candida sake, Hanseniaspora uvarum, and Kazachstania servazzii. Using a culture-dependent method, 41 strains representing the five major OTUs were isolated from the surface of the food samples. Whole genomes of the five major yeast strains were sequenced and annotated. The total genome length for the strains ranged from 8.97 Mbp to 21.32 Mbp. This is the first study to report genome sequences of the two yeasts Pichia kluyveri and Candida sake. Genome analysis indicated that each yeast strain had core metabolic pathways such as oxidative phosphorylation; purine metabolism; glycolysis/gluconeogenesis; aminoacyl-tRNA biosynthesis; citrate cycle; but strain specific pathways were also found. In addition, no toxin or antimicrobial resistance genes were identified. Our study provides genome information for five WCFY strains that may highlight their potential beneficial or harmful metabolic effects in fermented vegetables.
Similar content being viewed by others
References
Albertin, W., Setati, M.E., Miot-Sertier, C., Mostert, T.T., Colonna-Ceccaldi, B., Coulon, J., Girard, P., Moine, V., Pillet, M., Salin, F., et al. 2015. Hanseniaspora uvarum from winemaking environments show spatial and temporal genetic clustering. Front. Microbiol. 6, 1569.
Amara, A.A. and Shibl, A. 2015. Role of probiotics in health improvement, infection control and disease treatment and management. Saudi Pharm. J. 23, 107–114.
Barth, G. and Gaillardin, C. 1997. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol. Rev. 19, 219–237.
Bellemain, E., Carlsen, T., Brochmann, C., Coissac, E., Taberlet, P., and Kauserud, H. 2010. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiology 10, 189.
Buhagiar, R.W. and Barnett, J.A. 1971. The yeasts of strawberries. J. Appl. Bacteriol. 34, 727–739.
Cantarel, B.L., Korf, I., Robb, S.M., Parra, G., Ross, E., Moore, B., Holt, C., Sanchez Alvarado, A., and Yandell, M. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196.
Capece, A., Fiore, C., Maraz, A., and Romano, P. 2005. Molecular and technological approaches to evaluate strain biodiversity in Hanseniaspora uvarum of wine origin. J. Appl. Microbiol. 98, 136–144.
Carrau, F., Gaggero, C., and Aguilar, P.S. 2015. Yeast diversity and native vigor for flavor phenotypes. Trends Biotechnol. 33, 148–154.
Francisco, J., Narváez-Zapata, J.A., and Larralde-Corona, C.P. 2017. Microbial diversity and flavor quality of fermented beverages, pp. 125–154 In Microbial production of food ingredients and additives. Elsevier.
Hittinger, C.T., Steele, J.L., and Ryder, D.S. 2018. Diverse yeasts for diverse fermented beverages and foods. Curr. Opin. Biotechnol. 49, 199–206.
Jung, J.Y., Lee, S.H., and Jeon, C.O. 2014. Kimchi microflora: history, current status, and perspectives for industrial kimchi production. Appl. Microbiol. Biotechnol. 98, 2385–2393.
Kontis, V., Bennett, J.E., Mathers, C.D., Li, G., Foreman, K., and Ezzati, M. 2017. Future life expectancy in 35 industrialised countries: projections with a Bayesian model ensemble. Lancet 389, 1323–1335.
Kumar, S., Stecher, G., and Tamura, K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874.
Kurtzman, C.P. and Robnett, C.J. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73, 331–371.
Lanciotti, R., Gianotti, A., Baldi, D., Angrisani, R., Suzzi, G., Mastrocola, D., and Guerzoni, M. 2005. Use of Yarrowia lipolytica strains for the treatment of olive mill wastewater. Bioresour. Technol. 96, 317–322.
Letunic, I., Yamada, T., Kanehisa, M., and Bork, P. 2008. iPath: interactive exploration of biochemical pathways and networks. Trends Biochem. Sci. 33, 101–103.
Lu, Y., Voon, M.K.W., Chua, J.Y., Huang, D., Lee, P.R., and Liu, S.Q. 2017. The effects of co-and sequential inoculation of Torulaspora delbrueckii and Pichia kluyveri on chemical compositions of durian wine. Appl. Microbiol. Biotechnol. 101, 7853–7863.
Marco, M.L., Heeney, D., Binda, S., Cifelli, C.J., Cotter, P.D., Foligne, B., Gänzle, M., Kort, R., Pasin, G., and Pihlanto, A. 2017. Health benefits of fermented foods: microbiota and beyond. Curr. Opin. Biotechnol. 44, 94–102.
Marsh, A.J., Hill, C., Ross, R.P., and Cotter, P.D. 2014. Fermented beverages with health-promoting potential: past and future perspectives. Trends Food Sci. Technol. 38, 113–124.
Mei, J., Guo, Q., Wu, Y., and Li, Y. 2014. Microbial diversity of a camembert-type cheese using freeze-dried Tibetan kefir coculture as starter culture by culture-dependent and culture-independent methods. PLoS One 9, e111648.
Membré, J.M., Kubaczka, M., and Chéné, C. 1999. Combined effects of pH and sugar on growth rate of Zygosaccharomyces rouxii, a bakery product spoilage yeast. Appl. Environ. Microbiol. 65, 4921–4925.
Moon, S.H., Chang, M., Kim, H.Y., and Chang, H.C. 2014. Pichia kudriavzevii is the major yeast involved in film-formation, offodor production, and texture-softening in over-ripened Kimchi. Food Sci. Biotechnol. 23, 489–497.
Mukherjee, S., Stamatis, D., Bertsch, J., Ovchinnikova, G., Verezemska, O., Isbandi, M., Thomas, A.D., Ali, R., Sharma, K., Kyrpides, N.C., et al. 2017. Genomes online database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 45, D446–D456.
Nout, M.J.R. and Aidoo, K.E. 2002. Asian fungal fermented food, pp. 23–47. In Industrial applications. Springer.
Oh, Y.J., Jang, J.Y., Lim, S.K., Kwon, M.S., Lee, J., Kim, N., Shin, M.Y., Park, H.K., Seo, M.J., and Choi, H.J. 2017. Virgibacillus kimchii sp. nov., a halophilic bacterium isolated from kimchi. J. Microbiol. 55, 933–938.
Papanikolaou, S. and Aggelis, G. 2002. Lipid production by Yarrowia lipolytica growing on industrial glycerol in a single-stage continuous culture. Bioresour. Technol. 82, 43–49.
Polonelli, L., Menozzi, M.G., Campani, L., Gerloni, M., Conti, S., Morace, G., and Chezzi, C. 1992. Anaerobic yeast killer systems. Eur. J. Epidemiol. 8, 471–476.
Quirós, M., Rojas, V., Gonzalez, R., and Morales, P. 2014. Selection of non-Saccharomyces yeast strains for reducing alcohol levels in wine by sugar respiration. Int. J. Food Microbiol. 181, 85–91.
Rima, H., Steve, L., and Ismail, F. 2012. Antimicrobial and probiotic properties of yeasts: from fundamental to novel applications. Front. Microbiol. 3, 421.
Roh, S.W., Kim, K.H., Nam, Y.D., Chang, H.W., Park, E.J., and Bae, J.W. 2010. Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME J. 4, 1–16.
Romano, P., Fiore, C., Paraggio, M., Caruso, M., and Capece, A. 2003. Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 86, 169–180.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Spanoghe, M., Godoy Jara, M., Riviere, J., Lanterbecq, D., Gadenne, M., and Marique, T. 2017. Development and application of a quantitative real-time PCR assay for rapid detection of the multifaceted yeast Kazachstania servazzii in food. Food Microbiol. 62, 133–140.
Suzuki, A., Muraoka, N., Nakamura, M., Yanagisawa, Y., and Amachi, S. 2018. Identification of undesirable white-colony-forming yeasts appeared on the surface of Japanese kimchi. Biosci. Biotechnol. Biochem. 82, 334–342.
Tai, M. and Stephanopoulos, G. 2013. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 15, 1–9.
Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680.
Tominaga, T. 2004. Rapid identification of pickle yeasts by fluorescent PCR and microtemperature-gradient gel electrophoresis. FEMS Microbiol. Lett. 238, 43–48.
Vinas, I., Usall, J., Teixidó, N., and Sanchis, V. 1998. Biological control of major postharvest pathogens on apple with Candida sake. Int. J. Food Microbiol. 40, 9–16.
Walker, B.J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., Cuomo, C.A., Zeng, Q., Wortman, J., Young, S.K., et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963.
Yang, F., Hanna, M.A., and Sun, R. 2012. Value-added uses for crude glycerol--a byproduct of biodiesel production. Biotechnol. Biofuels 5, 13.
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., Aarestrup, F.M., and Larsen, M.V. 2012. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplemental material for this article may be found at https://doi.org/www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kim, J.Y., Kim, J., Cha, IT. et al. Community structures and genomic features of undesirable white colony-forming yeasts on fermented vegetables. J Microbiol. 57, 30–37 (2019). https://doi.org/10.1007/s12275-019-8487-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-019-8487-y