Abstract
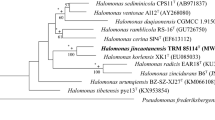
Strains pyc13T and ZGT13 were isolated from Lake Pengyan and Lake Zigetang on Tibetan Plateau, respectively. Both strains were Gram-negative, catalase- and oxidase-positive, aerobic, rod-shaped, nonmotile, and nonflagellated bacteria. Phylogenetic analysis based on 16S rRNA gene sequences showed that strains pyc13T and ZGT13 belong to the genus Halomonas, with Halomonas alkalicola 56-L4-10aEnT as their closest neighbor, showing 97.4% 16S rRNA gene sequence similarity. The predominant respiratory quinone of both strains was Q-9, with Q-8 as a minor component. The major fatty acids of both strains were C18:1ω6c/C18:1ω7c, C16:1ω6c/C16:1ω7c, C16:0, and C12:0 3OH. The polar lipids of both strains consisted of phosphatidylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, glycolipid, phospholipids of unknown structure containing glucosamine, and unidentified phospholipids. The DNA G + C content of pyc13T and ZGT13 were 62.6 and 63.4 mol%, respectively. The DNA-DNA hybridization values of strain pyc13T were 34, 41, 61, 35, and 35% with the reference strains H. alkalicola 56-L4-10aEnT, H. sediminicola CPS11T, H. mongoliensis Z-7009T, H. ventosae Al12T, and H. fontilapidosi 5CRT, respectively. Phenotypic, biochemical, genotypic, and DNA-DNA hybridization data showed that strains pyc13T and ZGT13 represent a new species within the genus Halomonas, for which the name H. tibetensis sp. nov. is proposed. The type strain is pyc13T (= CGMCC 1.15949T = KCTC 52660T).
Similar content being viewed by others
References
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990 Basic local alignment search tool. J. Mol. Biol. 215, 403–410
Arahal, D.R., Vreeland, R.H., Litchfield, C.D., Mormile, M.R., Tindall, B.J., Oren, A., Bejar, V., Quesada, E., and Ventosa, A. 2007 Recommended minimal standards for describing new taxa of the family Halomonadaceae. Int. J. Syst. Evol. Microbiol. 57, 2436–2446
Boltyanskaya, Y.V., Kevbrin, V.V., Lysenko, A.M., Kolganova, T.V., Tourova, T.P., Osipov, G.A., and Zhilina, T.N. 2007 Halomonas mongoliensis sp. nov. and Halomonas kenyensis sp. nov., new haloalkaliphilic denitrifiers capable of N2O reduction, isolated from soda lakes. Microbiology 76, 739–747.
de la Haba, R.R., Marquez, M.C., Papke, R.T., and Ventosa, A. 2012 Multilocus sequence analysis of the family Halomonadaceae. Int. J. Syst. Evol. Microbiol. 62, 520–538
De Ley, J., Cattoir, H., and Reynaerts, A. 1970 The quantitative measurement of DNA hybridization from renaturation rates. Eur. J. Biochem. 12, 133–142
Dobson, S.J. and Franzmann, P.D. 1996 Unification of the genera Deleya (Baumann et al. 1983), Halomonas (Vreeland et al. 1980), and Halovibrio (Fendric. 1988) and the species Paracoccus halodenitrificans (Robinson and Gibbon. 1952) into a single genus, Halomonas, and placement of the genus Zymobacter in the family Halomonadaceae. Int. J. Syst. Bacteriol. 46, 550–558
Dong, X.Z. and Cai, M.Y. 2001 Determinative manual for routine bacteriology. Beijing Scientific Press, Beijing, China.
Felsenstein, J. 1981 Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376
Gonzalez-Domenech, C.M., Martinez-Checa, F., Quesada, E., and Bejar, V. 2009 Halomonas fontilapidosi sp. nov., a moderately halophilic, denitrifying bacterium. Int. J. Syst. Evol. Microbiol. 59, 1290–1296
Guzman, D., Quillaguaman, J., Munoz, M., and Hatti-Kaul, R. 2010 Halomonas andesensis sp. nov., a moderate halophile isolated from the saline lake Laguna Colorada in Bolivia. Int. J. Syst. Evol. Microbiol. 60, 749–753
Heyrman, J. 2002 Halomonas muralis sp. nov., isolated from microbial biofilms colonizing the walls and murals of the Saint-Catherine chapel (Castle Herberstein, Austria). Int. J. Syst. Evol. Microbiol. 52, 2049–2054
Kaye, J.Z., Marquez, M.C., Ventosa, A., and Baross, J.A. 2004 Halomonas neptunia sp. nov., Halomonas sulfidaeris sp. nov., Halomonas axialensis sp. nov. and Halomonas hydrothermalis sp. nov.: halophilic bacteria isolated from deep-sea hydrothermal-vent environments. Int. J. Syst. Evol. Microbiol. 54, 499–511
Kim, K.K., Lee, K.C., Oh, H.M., and Lee, J.S. 2010 Halomonas stevensii sp. nov., Halomonas hamiltonii sp. nov. and Halomonas johnsoniae sp. nov., isolated from a renal care centre. Int. J. Syst. Evol. Microbiol. 60, 369–377
Kimura, M. 1979 The neutral theory of molecular evolution. Sci. Am. 241, 98–100, 102, 108
Kluge, A.G. and Farris, J.S. 1969 Quantitative phyletics and the evolution of Anurans. Syst. Zool. 18, 1–32
Kuykendall, L.D., Roy, M.A., O'Neill, J.J., and Devine, T.E. 1988 Fatty acids, antibiotic resistance and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int. J. Syst. Bacteriol. 38, 358–361
Lane, D.J. 1991. 16S/23S rRNA sequencing, pp. 115–175 In Stackebrandt, E. and Goodfellow, M. (eds.), Nucleic acid sequencing techniques in bacterial systematics, Wiley, New York, USA.
Lee, J.C., Kim, S.J., and Whang, K.S. 2016 Halomonas sediminicola sp. nov., a moderately halophilic bacterium isolated from a solar saltern sediment. Int. J. Syst. Evol. Microbiol. 66, 3865–3872
Marmur, J. and Doty, P. 1962 Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 5, 109–118
Martinez-Canovas, M.J., Quesada, E., Llamas, I., and Bejar, V. 2004 Halomonas ventosae sp. nov., a moderately halophilic, denitrifying, exopolysaccharide-producing bacterium. Int. J. Syst. Evol. Microbiol. 54, 733–737
Mata, J.A., Martinez-Canovas, J., Quesada, E., and Bejar, V. 2002 A detailed phenotypic characterisation of the type strains of Halomonas species. Syst. Appl. Microbiol. 25, 360–375
Minnikin, D.E., O'Donnell, A.G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., and Parlett, J.H. 1984 An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241
Qu, L., Lai, Q., Zhu, F., Hong, X., Zhang, J., Shao, Z., and Sun, X. 2011 Halomonas daqiaonensis sp. nov., a moderately halophilic, denitrifying bacterium isolated from a littoral saltern. Int. J. Syst. Evol. Microbiol. 61, 1612–1616
Quillaguaman, J., Hatti-Kaul, R., Mattiasson, B., Alvarez, M.T., and Delgado, O. 2004 Halomonas boliviensis sp. nov., an alkalitolerant, moderate halophile isolated from soil around a Bolivian hypersaline lake. Int. J. Syst. Evol. Microbiol. 54, 721–725
Saitou, N. and Nei, M. 1987 The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425
Sasser, M. 1990 Identification of bacteria through fatty acid analysis, pp. 199–204 In Klement, Z., Rudolph, K., and Sands, D.C. (eds.), Methods in Phytobacteriology, Akademiai Kaido, Budapest, Hungary.
Sehgal, S.N. and Gibbons, N.E. 1960 Effect of some metal ions on the growth of Halobacterium cutirubrum. Can. J. Microbiol. 6, 165–169
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. 2013 MEGA6: molecular evolutionary genetics analysis version 6.0 Mol. Biol. Evol. 30, 2725–2729
Tang, X., Zhai, L., Lin, Y., Yao, S., Wang, L., Ge, Y., Liu, Y., Zhang, X., Zhang, T., Zhang, L., et al. 2017 Halomonas alkalicola sp. nov., isolated from a household product plant. Int. J. Syst. Evol. Microbiol. 67, 1546–1550
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997 The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882
Tindall, B.J. 1990 Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol. Lett. 66, 199–202
Vreeland, R.H., Litchfield, C.D., Martin, E.L., and Elliot, E. 1980 Halomonas elongata, a new genus and species of extremely salttolerant bacteria. Int. J. Syst. Bacteriol. 30, 485–495
Wang, C.Y., Wu, S.J., Ng, C.C., Tzeng, W.S., and Shyu, Y.T. 2012 Halomonas beimenensis sp. nov., isolated from an abandoned saltern. Int. J. Syst. Evol. Microbiol. 62, 3013–3017
Wayne, L.G., Brenner, D.J., Colwell, R.R., Grimont, P.A.D., Kandler, O., Krichevsky, M.I., Moore, L.H., Moore, W.E.C., Murry, R.G.E., Stackebrandt, E., et al. 1987 Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464
Wu, Y.H., Xu, X.W., Huo, Y.Y., Zhou, P., Zhu, X.F., Zhang, H.B., and Wu, M. 2008 Halomonas caseinilytica sp. nov., a halophilic bacterium isolated from a saline lake on the Qinghai-Tibet Plateau, China. Int. J. Syst. Evol. Microbiol. 58, 1259–1262
Yoon, S.H., Ha, S.M., Kwon, S., Lim, J., Kim, Y., Seo, H., and Chun, J. 2016 Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617
Zhong, Z.P., Liu, Y., Wang, F., Zhou, Y.G., Liu, H.C., and Liu, Z.P. 2016 Lacimicrobium alkaliphilum gen. nov., sp. nov., a member of the family Alteromonadaceae isolated from a salt lake. Int. J. Syst. Evol. Microbiol. 66, 422–429
Author information
Authors and Affiliations
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Lu, Hb., Xing, P., Zhai, L. et al. Halomonas tibetensis sp. nov., isolated from saline lakes on Tibetan Plateau. J Microbiol. 56, 493–499 (2018). https://doi.org/10.1007/s12275-018-8076-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-018-8076-5