Abstract
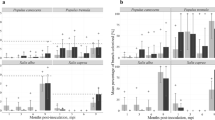
This study details the introduction of a gfp marker into an endophytic bacterial strain (Achromobacter marplatensis strain 17, isolated from sugar beet) to monitor its colonization of sugar beet (Beta. vulgaris L.). Stability of the plasmid encoding the gfp was confirmed in vitro for at least 72 h of bacterial growth and after the colonization of tissues, under nonselective conditions. The colonization was observed using fluorescence microscopy and enumeration of culturable endophytes in inoculated sugar beet plants that grew for 10 or 20 days. gfp-Expressing strains were re-isolated from the inner tissues of surface-sterilized roots and stems of inoculated plants, and the survival of the Achromobacter marplatensis 17:gfp strain in plants 20 days after inoculation, even in the absence of selective pressure, suggests that it is good colonizer. These results also suggest that this strain could be a useful tool for the delivery of enzymes or other proteins into plants. In addition, the study highlights that sugar beet plants can be used effectively for detailed in vitro studies on the interactions between A. marplatensis strain 17 and its host, particularly if a gfp-tagged strain of the pathogen is used.
Similar content being viewed by others
References
Andreote, F.D., Lacava, P.T., Gai, C.S., Araújo, W.L., Maccheroni, W.Jr., van Overbeek, L.S., van Elsas, J.D., and Azevedo, J.L. 2006. Model plants for studying the interaction between Methylobacterium mesophilicum and Xylella fastidiosa. Can. J. Microbiol. 52, 419–426.
Araújo, W.L., Maccheroni, W. Jr., Aguilar-Vildoso, C.I., Barroso, P.A., Saridakis, H.O., and Azevedo, J.L. 2001. Variability and interactions between endophytic bacteria and fungi isolated from leaf tissues of citrus rootstocks. Can. J. Microbiol. 47, 229–236.
Chi, F., Shen, S.H., Cheng, H.P., Jing, Y.X., Yanni, Y.G., and Dazzo, F.B. 2005. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl. Environ. Microbiol. 71, 7271–7278.
Compant, S., Reiter, B., Sessitsch, A., Nowak, J., Clement, C., and Barka, E.A. 2005. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 71, 1685–1693.
Cormack, B.P., Valdivia, R.H., and Falkow, S. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38.
Dong, Y., Iniguez, A.L., and Triplett, E.W. 2003. Quantitative assessments of the host range and strain specifcity of endophytic colonization by Klebsiella pneumoniae 342. Plant Soil 257, 49–59.
Errampalli, D., Leung, K., Cassidy, M.B., Kostrzynska, M., Blears, M., Lee, H., and Trevors, J.T. 1999. Applications of the green fluorescent protein as a molecular marker in environmental microorganisms. J. Microbiol. Methods 35, 187–199.
Ferreira, A., Quecine, M.C., Lacava, P.T., Oda, S., Azevedo, J.L., and Araújo, W.L. 2008. Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol. Lett. 287, 8–14.
Germaine, K., Keogh, E., Garcia-Cabellos, G., Borremans, B., van der Lelie, D., Barac, T., Oeyen, L., Vangronsveld, J., Moore, P.F., Moore, E.R.B., et al. 2004. Colonisation of poplar trees by gfp expressing bacterial endophytes. FEMS Microbiol. Ecol. 48, 109–118.
Glick, B.R. 1995. The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 41, 109–117.
Haas, D. and Défago, G. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319.
Jacobs, M.J., William, M.B., and David, A.G. 1985. Enumeration, location, characterization of endophytic bacteria within sugar beet roots. Can. J. Bot. 63, 1262–1265.
Kuklinsky-Sobral, J., Araujo, W.L., Mendes, R., Pizzirani-Kleiner, A.A., and Azevedo, J.L. 2005. Isolation and characterization of endophytic bacteria from soybean (Glycine max) grown in soil treated with glyphosate herbicide. Plant Soil 273, 91–99.
Lacava, P.T., Li, W., Araujo, W.L., Azevedo, J.L., and Hartung, J.S. 2007. The endophyte Curtobacterium flaccumfaciens reduces symptoms caused by Xylella fastidiosa in Catharanthus roseus. J. Microbiol. 45, 388–393.
Lodewyck, C., Vangronsveld, J., Porteous, F., Moore, E.R.B., Taghavi, S., Mezgeay, M., and van der Lelie, D. 2002. Endophytic bacteria and their potential application. Crit. Rev. Plant Sci. 86, 583–606.
Lucas-Garca, J.A., Schloter, M., Durkaya, T., Hartmann, A., and Gutirrez-Maero, F.J. 2003. Colonization of pepper roots by a plant growth promoting Pseudomonas fluorescens strain. Biol. Fertil. Soils 37, 381–385.
Mareque, C., Taule, C., Beracochea, M., and Battistoni, F. 2015. Isolation, characterization and plant growth promotion effects of putative bacterial endophytes associated with sweet sorghum (Sorghum bicolor (L) Moench). Ann. Microbiol. 65, 1057–1067.
Mcinroy, J.A. and Kloepper, J.W. 1995. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant Soil 173, 337–342.
Nairn, J.D. and Chanway, C.P. 2002. Temporary loss of antibiotic resistance by marked bacteria in the rhizosphere of spruce seedlings. FEMS Microbiol. Ecol. 40, 167–170.
Newman, L.A. and Reynolds, C.M. 2005. Bacteria and phytoremediation: new uses for endophytic bacteria in plants. Trends Biotechnol. 23, 6–8.
Qazi, S.N.A., Rees, C.E.D., Mellits, K.H., and Hill, P.J. 2001. Development of gfp vectors for expression in Listeria monocytogenes and other low G+C gram positive bacteria. Microb. Ecol. 41, 301–309.
Ryan, R.P., Germaine, K., Franks, A., Ryan, D.J., and Dowling, D.N. 2008. Bacterial endophytes: recent developments and applications. FEMS Microbiol. Lett. 278, 1–9.
Shi, Y.W., Gao, Y., Yang, H.M., Zhang, T., Lin, Q., Chang, W., Li, Y.G., Outikuer, M., and Lou, K. 2014. A beet endogenous growth promoting bacteria and its application. Xinjiang: CN103642733A, 2014-03-19.
Shi, Y., Lou, K., and Li, C. 2008. Growth promotion effects of the endophyte Acinetobacter johnsonii strain 3-1 on sugar beet. Symbiosis 54, 159–166.
Tian, T., Qi, X.C., Wang, Q., and Mei, R.H. 2004 Colonization study of GFP-tagged Bacillus strains on wheat surface. Acta Phytopathol. Sin. 34, 346–351.
Valdivia, R.H. and Falkow, S. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277, 2007–2011.
Verma, S.C., Singh, A., Chowdhury, S.P., and Tripathi, A.K. 2004. Endophytic colonization ability of two deep-water rice endophytes, Pantoea sp. and Ochrobactrum sp. using green fluorescent protein reporter. Biotechnol. Lett. 26, 425–429.
Wu, Z., Peng, Y., Guo, L., and Li, C. 2014. Root colonization of encapsulated Klebsiella oxytoca Rs-5 on cotton plants and its promoting growth performance under salinity stress. European J. Soil Biol. 60, 81–87.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shi, Y., Li, C., Yang, H. et al. Colonization study of gfp-tagged Achromobacter marplatensis strain in sugar beet. J Microbiol. 55, 267–272 (2017). https://doi.org/10.1007/s12275-017-6371-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-017-6371-1