Abstract
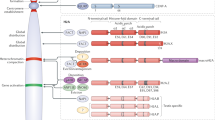
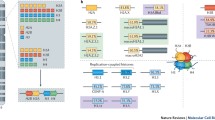
Chromatin is one of the most significant structures to regulate gene expression. Active deposition and incorporation of specialized histone variants can lead to severe alterations in gene expression, cell cycle progression and embryonic development. Further, deregulation or mutations of histone variants are often implicated in diseases, most notably cancer development. Our work focuses on the discovery of novel human histone variants and variant-interactors to better understand fundamental epigenetic processes.
Similar content being viewed by others
Literatur
Goldberg AD, Allis CD, Bernstein E (2007) Epigenetics: a landscape takes shape. Cell 128:635–638
Chittka A, Chittka L (2010) Epigenetics of royality. PLoS Biol 8:e1000532
Vardabasso C, Gaspar-Maia A, Hasson D et al. (2015) Histone variant H2A.Z.2 mediates proliferation and drug sensitivity of malignant melanoma. Mol Cell 59:75–88
Bönisch C, Hake SB (2012) Histone variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Res 40:10719–10741
Bönisch C, Nieratschker SM, Orfanos NK et al. (2008) Chromatin proteomics and epigenetic regulatory circuits. Expert Rev Proteomics 5:105–119
Wiedemann SM, Mildner SN, Bönisch C et al. (2010) Identification and characterization of two novel primatespecific histone H3 variants, H3.X and H3.Y. J Cell Biol 190:777–791
Zink LM, Delbarre E, Eberl HC et al. (2017) H3.Y discriminates between HIRA and DAXX chaperone complexes and reveals unexpected insights into human DAXX-H3.3-H4 binding and deposition requirements. Nucleic Acids Res 45:5691–5706
Bönisch C, Schneider K, Pünzeler S et al. (2012) H2A.Z.2.2 is an alternatively spliced histone H2A.Z variant that causes severe nucleosome destabilization. Nucleic Acids Res 40:5951–5964
Pünzeler S, Link S, Wagner G et al. (2017) Multivalent binding of PWWP2A to H2A.Z regulates mitosis and neural crest differentiation. EMBO J 36:2263–2279
Zink LM, Hake SB (2016) Histone variants: nuclear function and disease. Curr Opin Genet Dev 37:82–89
Author information
Authors and Affiliations
Corresponding authors
Additional information
Ramona Spitzer 2008–2012 Bachelorstudium Molekulare Biotechnologie und 2012–2015 Masterstudium Biologie an der TU München. Seit 2015 Promotion in der Abteilung für Molekularbiologie an der LMU München.
Sandra Hake Biologiestudium an der RWTH Aachen und Promotion am Max-Planck-Institut für Immunbiologie, Freiburg. 2000–2003 Postdoc am Memorial-Sloan-Kettering Cancer Institute, New York, USA, und 2003–2006 an der Rockefeller University, New York. 2006–2016 Gruppenleiterin in der Abteilung für Molekularbiologie an der LMU München. Seit 2016 Profeßsorin (W3) für Genetik an der Universität Gießen.
Rights and permissions
About this article
Cite this article
Spitzer, R.M.M., Hake, S.B. Histonvarianten - Gleiche Gene bedeuten nicht gleiches Schicksal. Biospektrum 23, 752–755 (2017). https://doi.org/10.1007/s12268-017-0865-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12268-017-0865-6