Abstract
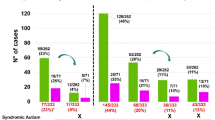
Chromosome microarray analysis (CMA) is a cost-effective molecular cytogenetic technique that has been used as a first-line diagnostic test in neurodevelopmental disorders in the USA since 2011. The impact of CMA results on clinical practice in China is not yet well studied, so we aimed to better evaluate this phenomenon. We analyzed the CMA results from 434 patients in our clinic, and characterized their molecular diagnoses, clinical features, and follow-up clinical actions based on these results. The overall diagnostic yield for our patients was 13.6% (59 out of 434). This gave a detection rate of 14.7% for developmental delay/intellectual disability (DD/ID, 38/259) and 12% for autism spectrum disorders (ASDs, 21/175). Thirty-three recurrent (n ≥ 2) variants were found, distributed at six chromosomal loci involving known chromosome syndromes (such as DiGeorge, Williams Beuren, and Angelman/Prader-Willi syndromes). The spectrum of positive copy number variants in our study was comparable to that reported in Caucasian populations, but with specific characteristics. Parental origin tests indicated an effect involving a significant maternal transmission bias to sons. The majority of patients with positive results (94.9%) had benefits, allowing earlier diagnosis (36/59), prioritized full clinical management (28/59), medication changes (7/59), a changed prognosis (30/59), and prenatal genetic counseling (15/59). Our results provide information on de novo mutations in Chinese children with DD/ID and/or ASDs. Our data showed that microarray testing provides immediate clinical utility for patients. It is expected that the personalized medical care of children with developmental disabilities will lead to improved outcomes in long-term developmental potential. We advocate using the diagnostic yield of clinically actionable results to evaluate CMA as it provides information of both clinical validity and clinical utility.

Similar content being viewed by others
References
Boyle CA, Boulet S, Schieve LA, Cohen RA, Blumberg SJ, Yeargin-Allsopp M, et al. Trends in the prevalence of developmental disabilities in US children, 1997-2008. Pediatrics 2011, 127: 1034–1042.
Michelson DJ, Shevell MI, Sherr EH, Moeschler JB, Gropman AL, Ashwal S. Evidence report: Genetic and metabolic testing on children with global developmental delay: report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology 2011, 77: 1629–1635.
Christensen DL, Baio J, Van Naarden Braun K, Bilder D, Charles J, Constantino JN, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years–Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2012. MMWR Surveill Summ 2016, 65: 1–23.
Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010, 86: 749–764.
Manning M, Hudgins L. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med 2010, 12: 742–745.
Blue Cross Blue Shield Asssociation. Special report: Chromosomal microarray for the genetic evaluation of patients with global developmental delay, intellectual disability, and autism spectrum disorder. Technol Eval Cent Assess Program Exec Summ 2015, 30: 1–4.
Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 2008, 82: 477–488.
Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 2010, 466: 368–372.
Coe BP, Witherspoon K, Rosenfeld JA, van Bon BW, Vulto-van Silfhout AT, Bosco P, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet 2014, 46: 1063–1071.
Coulter ME, Miller DT, Harris DJ, Hawley P, Picker J, Roberts AE, et al. Chromosomal microarray testing influences medical management. Genet Med 2011, 13: 770–776.
Riggs ER, Wain KE, Riethmaier D, Smith-Packard B, Faucett WA, Hoppman N, et al. Chromosomal microarray impacts clinical management. Clin Genet 2014, 85: 147–153.
Henderson LB, Applegate CD, Wohler E, Sheridan MB, Hoover-Fong J, Batista DA. The impact of chromosomal microarray on clinical management: a retrospective analysis. Genet Med 2014, 16: 657–664.
Tao VQ, Chan KY, Chu YW, Mok GT, Tan TY, Yang W, et al. The clinical impact of chromosomal microarray on paediatric care in Hong Kong. PLoS One 2014, 9: e109629.
Yin CL, Chen HI, Li LH, Chien YL, Liao HM, Chou MC, et al. Genome-wide analysis of copy number variations identifies PARK2 as a candidate gene for autism spectrum disorder. Mol Autism 2016, 7: 23.
Gazzellone MJ, Zhou X, Lionel AC, Uddin M, Thiruvahindrapuram B, Liang S, et al. Copy number variation in Han Chinese individuals with autism spectrum disorder. J Neurodev Disord 2014, 6: 34.
Chong WW, Lo IF, Lam ST, Wang CC, Luk HM, Leung TY, et al. Performance of chromosomal microarray for patients with intellectual disabilities/developmental delay, autism, and multiple congenital anomalies in a Chinese cohort. Mol Cytogenet 2014, 7: 34.
Wang B, Ji T, Zhou X, Wang J, Wang X, Wang J, et al. CNV analysis in Chinese children of mental retardation highlights a sex differentiation in parental contribution to de novo and inherited mutational burdens. Sci Rep 2016, 6: 25954.
Harfterkamp M, Buitelaar JK, Minderaa RB, van de Loo-Neus G, van der Gaag RJ, Hoekstra PJ. Long-term treatment with atomoxetine for attention-deficit/hyperactivity disorder symptoms in children and adolescents with autism spectrum disorder: an open-label extension study. J Child Adolesc Psychopharmacol 2013, 23: 194–199.
Sagoo GS, Mohammed S, Barton G, Norbury G, Ahn JW, Ogilvie CM, et al. Cost effectiveness of using array-CGH for diagnosing learning disability. Appl Health Econ Health Policy 2015, 13: 421–432.
Trakadis Y, Shevell M. Microarray as a first genetic test in global developmental delay: a cost-effectiveness analysis. Dev Med Child Neurol 2011, 53: 994–999.
Morrow EM. Genomic copy number variation in disorders of cognitive development. J Am Acad Child Adolesc Psychiatry 2010, 49: 1091–1104.
Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA 2015, 314: 895–903.
Xu LM, Li JR, Huang Y, Zhao M, Tang X, Wei L. AutismKB: an evidence-based knowledgebase of autism genetics. Nucleic Acids Res 2012, 40: D1016–1022.
Huang AX, Jia M, Wheeler JJ. Children with autism in the People’s Republic of China: diagnosis, legal issues, and educational services. J Autism Dev Disord 2013, 43: 1991–2001.
Zeng Y, Hesketh T. The effects of China’s universal two-child policy. Lancet 2016, 388: 1930–1938.
Duyzend MH, Nuttle X, Coe BP, Baker C, Nickerson DA, Bernier R, et al. Maternal modifiers and parent-of-origin bias of the autism-associated 16p11.2 CNV. Am J Hum Genet 2016, 98: 45–57.
Thomas NS, Durkie M, Potts G, Sandford R, Van Zyl B, Youings S, et al. Parental and chromosomal origins of microdeletion and duplication syndromes involving 7q11.23, 15q11-q13 and 22q11. Eur J Hum Genet 2006, 14: 831–837.
Delio M, Guo T, McDonald-McGinn DM, Zackai E, Herman S, Kaminetzky M, et al. Enhanced maternal origin of the 22q11.2 deletion in velocardiofacial and DiGeorge syndromes. Am J Hum Genet 2013, 92: 439–447.
Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet 2015, 47: 582–588.
Beaudet AL. Reaching a CNV milestone. Nat Genet 2014, 46: 1046–1048.
Shishido E, Aleksic B, Ozaki N. Copy-number variation in the pathogenesis of autism spectrum disorder. Psychiatry Clin Neurosci 2014, 68: 85–95.
Rosenfeld JA, Coppinger J, Bejjani BA, Girirajan S, Eichler EE, Shaffer LG, et al. Speech delays and behavioral problems are the predominant features in individuals with developmental delays and 16p11.2 microdeletions and microduplications. J Neurodev Disord 2010, 2: 26–38.
Anguera JA, Brandes-Aitken AN, Rolle CE, Skinner SN, Desai SS, Bower JD, et al. Characterizing cognitive control abilities in children with 16p11.2 deletion using adaptive ‘video game’ technology: a pilot study. Transl Psychiatry 2016, 6: e893.
Shinawi M, Liu P, Kang SH, Shen J, Belmont JW, Scott DA, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet 2010, 47: 332–341.
Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet 2008, 17: 628–638.
Wu N, Ming X, Xiao J, Wu Z, Chen X, Shinawi M, et al. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N Engl J Med 2015, 372: 341–350.
Abbas E, Cox DM, Smith T, Butler MG. The 7q11.23 microduplication syndrome: A clinical report with review of literature. J Pediatr Genet 2016, 5: 129–140.
Siu WK, Lam CW, Mak CM, Lau ET, Tang MH, Tang WF, et al. Diagnostic yield of array CGH in patients with autism spectrum disorder in Hong Kong. Clin Transl Med 2016, 5: 18.
Ke Q, Zhang L, He C, Zhao Z, Qi M, Griggs RC, et al. China’s shift from population control to population quality: Implications for neurology. Neurology 2016, 87: e85–88.
Subspecialty Group of Clinical Genetics, Society of Adolescent Medicine, Chinese Medical Doctor Association; Society of Medical Genetics, Chinese Medical Doctor Association; Subspecialty Group of Endocrinologic, Hereditary and Metabolic Diseases, The Society of Pediatrics, Chinese Medical Association. Expert consensus on the clinical application of chromosomal microarray analysis in pediatric genetic diseases. Chin J Pediatr 2016, 54: 410–413.
Acknowledgements
We thank all of the families who participated in this project. This work was supported by grants from the National Natural Science Foundation of China (81761128035 and 81781220701), the Shanghai Municipal Science and Technology Committee (17XD1403200 and 18dz2313505), the Research Physician Project of Shanghai Municipal Education Commission (20152234), the Shanghai Municipal Health and Family Planning Commission (GDEK201709, 2017ZZ02026, and 2017EKHWYX-02), and the Scientific Program of Shanghai Shenkang Hospital Development Center (16CR2025B) of China.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors claim that there are no conflicts of interest.
Rights and permissions
About this article
Cite this article
Xu, M., Ji, Y., Zhang, T. et al. Clinical Application of Chromosome Microarray Analysis in Han Chinese Children with Neurodevelopmental Disorders. Neurosci. Bull. 34, 981–991 (2018). https://doi.org/10.1007/s12264-018-0238-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12264-018-0238-2