Abstract
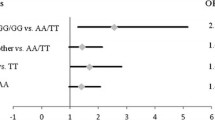
A growing number of studies have suggested that genetic variants affecting the micro-RNA- binding mechanisms (miRSNPs) constitute a promising novel class of biomarkers for prostate cancer (PCa) biology. Among the most extensively studied miRSNPs in the context of cancer is the variation rs4245739 in the MDM4 gene, while a recent large-scale analysis revealed significant differences in genotype distributions between aggressive and non-aggressive disease for rs1058205 in KLK3 and rs1010 in VAMP8. In this study, we examined a total of 1083 subjects for these three variants using Taqman® SNP Genotyping Assays. Three hundred and fifty-five samples of peripheral blood were obtained from patients with PCa and 358 samples from patients with benign prostatic hyperplasia (BPH). The control group consisted of 370 healthy volunteers. Comparisons of genotype distributions among PCa and BPH patients, as well as between PCa patients and healthy controls, yielded no evidence of association between the analyzed genetic variants and the risk of developing PCa. However, all three tested genetic variants have shown the association with the parameters of PCa progression. For KLK3 variant rs1058205, minor allele C was found to associate with the lower serum PSA score in PCa patients (PSA > 20 ng/ml vs. PSA < 10 ng/ml comparison, Prec = 0.038; ORrec = 0.20, 95%CI 0.04–1.05). The obtained results point out the potential relevance of the tested genetic variants for the disease aggressiveness assessment.
Similar content being viewed by others
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424. https://doi.org/10.3322/caac.21492
Eeles R, Ni Raghallaigh H (2018) Men with a susceptibility to prostate cancer and the role of genetic based screening. Translat Androl Urol 7(1):61–69. https://doi.org/10.21037/tau.2017.12.30
Schumacher FR, Al Olama AA, Berndt SI, Benlloch S, Ahmed M, Saunders EJ, Dadaev T, Leongamornlert D, Anokian E, Cieza-Borrella C, Goh C, Brook MN, Sheng X, Fachal L, Dennis J, Tyrer J, Muir K, Lophatananon A, Stevens VL, Gapstur SM, Carter BD, Tangen CM, Goodman PJ, Thompson IM Jr, Batra J, Chambers S, Moya L, Clements J, Horvath L, Tilley W, Risbridger GP, Gronberg H, Aly M, Nordstrom T, Pharoah P, Pashayan N, Schleutker J, Tammela TLJ, Sipeky C, Auvinen A, Albanes D, Weinstein S, Wolk A, Hakansson N, West CML, Dunning AM, Burnet N, Mucci LA, Giovannucci E, Andriole GL, Cussenot O, Cancel-Tassin G, Koutros S, Beane Freeman LE, Sorensen KD, Orntoft TF, Borre M, Maehle L, Grindedal EM, Neal DE, Donovan JL, Hamdy FC, Martin RM, Travis RC, Key TJ, Hamilton RJ, Fleshner NE, Finelli A, Ingles SA, Stern MC, Rosenstein BS, Kerns SL, Ostrer H, Lu YJ, Zhang HW, Feng N, Mao X, Guo X, Wang G, Sun Z, Giles GG, Southey MC, MacInnis RJ, FitzGerald LM, Kibel AS, Drake BF, Vega A, Gomez-Caamano A, Szulkin R, Eklund M, Kogevinas M, Llorca J, Castano-Vinyals G, Penney KL, Stampfer M, Park JY, Sellers TA, Lin HY, Stanford JL, Cybulski C, Wokolorczyk D, Lubinski J, Ostrander EA, Geybels MS, Nordestgaard BG, Nielsen SF, Weischer M, Bisbjerg R, Roder MA, Iversen P, Brenner H, Cuk K, Holleczek B, Maier C, Luedeke M, Schnoeller T, Kim J, Logothetis CJ, John EM, Teixeira MR, Paulo P, Cardoso M, Neuhausen SL, Steele L, Ding YC, De Ruyck K, De Meerleer G, Ost P, Razack A, Lim J, Teo SH, Lin DW, Newcomb LF, Lessel D, Gamulin M, Kulis T, Kaneva R, Usmani N, Singhal S, Slavov C, Mitev V, Parliament M, Claessens F, Joniau S, Van den Broeck T, Larkin S, Townsend PA, Aukim-Hastie C, Gago-Dominguez M, Castelao JE, Martinez ME, Roobol MJ, Jenster G, van Schaik RHN, Menegaux F, Truong T, Koudou YA, Xu J, Khaw KT, Cannon-Albright L, Pandha H, Michael A, Thibodeau SN, McDonnell SK, Schaid DJ, Lindstrom S, Turman C, Ma J, Hunter DJ, Riboli E, Siddiq A, Canzian F, Kolonel LN, Le Marchand L, Hoover RN, Machiela MJ, Cui Z, Kraft P, Amos CI, Conti DV, Easton DF, Wiklund F, Chanock SJ, Henderson BE, Kote-Jarai Z, Haiman CA, Eeles RA, Profile S, Australian Prostate Cancer B, Study I, Canary PI, Breast, Prostate Cancer Cohort C, Consortium P, Cancer of the Prostate in S, Prostate Cancer Genome-wide Association Study of Uncommon Susceptibility L, Genetic A, Mechanisms in Oncology /Elucidating Loci Involved in Prostate Cancer Susceptibility C (2018) Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet 50(7):928–936. https://doi.org/10.1038/s41588-018-0142-8
Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, Suveges D, Vrousgou O, Whetzel PL, Amode R, Guillen JA, Riat HS, Trevanion SJ, Hall P, Junkins H, Flicek P, Burdett T, Hindorff LA, Cunningham F, Parkinson H (2019) The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47(D1):D1005–D1012. https://doi.org/10.1093/nar/gky1120
Wilk G, Braun R (2018) regQTLs: single nucleotide polymorphisms that modulate microRNA regulation of gene expression in tumors. PLoS Genet 14(12):e1007837. https://doi.org/10.1371/journal.pgen.1007837
Vishnoi A, Rani S (2017) MiRNA biogenesis and regulation of diseases: an overview. Methods Mol Biol 1509:1–10. https://doi.org/10.1007/978-1-4939-6524-3_1
Salzman DW, Weidhaas JB (2013) SNPing cancer in the bud: microRNA and microRNA-target site polymorphisms as diagnostic and prognostic biomarkers in cancer. Pharmacol Ther 137(1):55–63. https://doi.org/10.1016/j.pharmthera.2012.08.016
Cipollini M, Landi S, Gemignani F (2014) MicroRNA binding site polymorphisms as biomarkers in cancer management and research. Pharmacogen Personal Med 7:173–191. https://doi.org/10.2147/PGPM.S61693
Yuan Y, Weidhaas JB (2019) Functional microRNA binding site variants. Mol Oncol 13(1):4–8. https://doi.org/10.1002/1878-0261.12421
Kotarac N, Dobrijevic Z, Matijasevic S, Savic-Pavicevic D, Brajuskovic G (2019) Analysis of association of potentially functional genetic variants within genes encoding miR-34b/c, miR-378 and miR-143/145 with prostate cancer in Serbian population. EXCLI J 18:515–529. https://doi.org/10.17179/excli2019-1257
Nikolic Z, Savic Pavicevic D, Vucic N, Cidilko S, Filipovic N, Cerovic S, Vukotic V, Romac S, Brajuskovic G (2015) Assessment of association between genetic variants in microRNA genes hsa-miR-499, hsa-miR-196a2 and hsa-miR-27a and prostate cancer risk in Serbian population. Exp Mol Pathol 99(1):145–150. https://doi.org/10.1016/j.yexmp.2015.06.009
Nikolic ZZ, Savic Pavicevic DL, Vukotic VD, Tomovic SM, Cerovic SJ, Filipovic N, Romac SP, Brajuskovic GN (2014) Association between genetic variant in hsa-miR-146a gene and prostate cancer progression: evidence from Serbian population. Cancer Causes Cont: CCC 25(11):1571–1575. https://doi.org/10.1007/s10552-014-0452-9
Hughes L, Ruth K, Rebbeck TR, Giri VN (2013) Genetic variation in IL-16 miRNA target site and time to prostate cancer diagnosis in African-American men. Prostate Cancer Prostatic Dis 16(4):308–314. https://doi.org/10.1038/pcan.2013.36
Liu J, Huang J, He Y, Liu J, Liao B, Liao G (2014) Genetic variants in the integrin gene predicted microRNA-binding sites were associated with the risk of prostate cancer. Mol Carcinog 53(4):280–285. https://doi.org/10.1002/mc.21973
Stegeman S, Amankwah E, Klein K, O'Mara TA, Kim D, Lin HY, Permuth-Wey J, Sellers TA, Srinivasan S, Eeles R, Easton D, Kote-Jarai Z, Amin Al Olama A, Benlloch S, Muir K, Giles GG, Wiklund F, Gronberg H, Haiman CA, Schleutker J, Nordestgaard BG, Travis RC, Neal D, Pharoah P, Khaw KT, Stanford JL, Blot WJ, Thibodeau S, Maier C, Kibel AS, Cybulski C, Cannon-Albright L, Brenner H, Kaneva R, Teixeira MR, Consortium P, Australian Prostate Cancer B, Spurdle AB, Clements JA, Park JY, Batra J (2015) A large-scale analysis of genetic variants within putative miRNA binding sites in Prostate Cancer. Cancer Discov 5(4):368–379. https://doi.org/10.1158/2159-8290.CD-14-1057
Stegeman S, Moya L, Selth LA, Spurdle AB, Clements JA, Batra J (2015) A genetic variant of MDM4 influences regulation by multiple microRNAs in prostate cancer. Endocr Relat Cancer 22(2):265–276. https://doi.org/10.1530/ERC-15-0013
Hong SK (2014) Kallikreins as biomarkers for prostate cancer. Biomed Res Int 2014:526341–526310. https://doi.org/10.1155/2014/526341
Bensen JT, Xu Z, Smith GJ, Mohler JL, Fontham ET, Taylor JA (2013) Genetic polymorphism and prostate cancer aggressiveness: a case-only study of 1,536 GWAS and candidate SNPs in African-Americans and European-Americans. Prostate 73(1):11–22. https://doi.org/10.1002/pros.22532
Savblom C, Hallden C, Cronin AM, Sall T, Savage C, Vertosick EA, Klein RJ, Giwercman A, Lilja H (2014) Genetic variation in KLK2 and KLK3 is associated with concentrations of hK2 and PSA in serum and seminal plasma in young men. Clin Chem 60(3):490–499. https://doi.org/10.1373/clinchem.2013.211219
Chen C, Xin Z (2017) Single-nucleotide polymorphism rs1058205 of KLK3 is associated with the risk of prostate cancer: a case-control study of Han Chinese men in Northeast China. Medicine 96(10):e6280. https://doi.org/10.1097/MD.0000000000006280
Penney KL, Schumacher FR, Kraft P, Mucci LA, Sesso HD, Ma J, Niu Y, Cheong JK, Hunter DJ, Stampfer MJ, Hsu SI (2011) Association of KLK3 (PSA) genetic variants with prostate cancer risk and PSA levels. Carcinogenesis 32(6):853–859. https://doi.org/10.1093/carcin/bgr050
Wang CC, Shi H, Guo K, Ng CP, Li J, Gan BQ, Chien Liew H, Leinonen J, Rajaniemi H, Zhou ZH, Zeng Q, Hong W (2007) VAMP8/endobrevin as a general vesicular SNARE for regulated exocytosis of the exocrine system. Mol Biol Cell 18(3):1056–1063. https://doi.org/10.1091/mbc.e06-10-0974
Zhu D, Zhang Y, Lam PP, Dolai S, Liu Y, Cai EP, Choi D, Schroer SA, Kang Y, Allister EM, Qin T, Wheeler MB, Wang CC, Hong WJ, Woo M, Gaisano HY (2012) Dual role of VAMP8 in regulating insulin exocytosis and islet beta cell growth. Cell Metab 16(2):238–249. https://doi.org/10.1016/j.cmet.2012.07.001
Gansmo LB, Bjornslett M, Halle MK, Salvesen HB, Dorum A, Birkeland E, Hveem K, Romundstad P, Vatten L, Lonning PE, Knappskog S (2016) The MDM4 SNP34091 (rs4245739) C-allele is associated with increased risk of ovarian-but not endometrial cancer. Tumour Biol 37(8):10697–10702. https://doi.org/10.1007/s13277-016-4940-2
Gao F, Xiong X, Pan W, Yang X, Zhou C, Yuan Q, Zhou L, Yang M (2015) A regulatory MDM4 genetic variant locating in the binding sequence of multiple MicroRNAs contributes to susceptibility of small cell lung Cancer. PLoS One 10(8):e0135647. https://doi.org/10.1371/journal.pone.0135647
Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, Orr N, Rhie SK, Riboli E, Feigelson HS, Le Marchand L, Buring JE, Eccles D, Miron P, Fasching PA, Brauch H, Chang-Claude J, Carpenter J, Godwin AK, Nevanlinna H, Giles GG, Cox A, Hopper JL, Bolla MK, Wang Q, Dennis J, Dicks E, Howat WJ, Schoof N, Bojesen SE, Lambrechts D, Broeks A, Andrulis IL, Guenel P, Burwinkel B, Sawyer EJ, Hollestelle A, Fletcher O, Winqvist R, Brenner H, Mannermaa A, Hamann U, Meindl A, Lindblom A, Zheng W, Devillee P, Goldberg MS, Lubinski J, Kristensen V, Swerdlow A, Anton-Culver H, Dork T, Muir K, Matsuo K, Wu AH, Radice P, Teo SH, Shu XO, Blot W, Kang D, Hartman M, Sangrajrang S, Shen CY, Southey MC, Park DJ, Hammet F, Stone J, Veer LJ, Rutgers EJ, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Peto J, Schrauder MG, Ekici AB, Beckmann MW, Dos Santos SI, Johnson N, Warren H, Tomlinson I, Kerin MJ, Miller N, Marme F, Schneeweiss A, Sohn C, Truong T, Laurent-Puig P, Kerbrat P, Nordestgaard BG, Nielsen SF, Flyger H, Milne RL, Perez JI, Menendez P, Muller H, Arndt V, Stegmaier C, Lichtner P, Lochmann M, Justenhoven C, Ko YD, Gene EI, breast CN, Muranen TA, Aittomaki K, Blomqvist C, Greco D, Heikkinen T, Ito H, Iwata H, Yatabe Y, Antonenkova NN, Margolin S, Kataja V, Kosma VM, Hartikainen JM, Balleine R, kConFab I, Tseng CC, Berg DV, Stram DO, Neven P, Dieudonne AS, Leunen K, Rudolph A, Nickels S, Flesch-Janys D, Peterlongo P, Peissel B, Bernard L, Olson JE, Wang X, Stevens K, Severi G, Baglietto L, MClean C, Coetzee GA, Feng Y, Henderson BE, Schumacher F, Bogdanova NV, Labreche F, Dumont M, Yip CH, Taib NA, Cheng CY, Shrubsole M, Long J, Pylkas K, Jukkola-Vuorinen A, Kauppila S, Knight JA, Glendon G, Mulligan AM, Tollenaar RA, Seynaeve CM, Kriege M, Hooning MJ, van den Ouweland AM, van Deurzen CH, Lu W, Gao YT, Cai H, Balasubramanian SP, Cross SS, Reed MW, Signorello L, Cai Q, Shah M, Miao H, Chan CW, Chia KS, Jakubowska A, Jaworska K, Durda K, Hsiung CN, Wu PE, Yu JC, Ashworth A, Jones M, Tessier DC, Gonzalez-Neira A, Pita G, Alonso MR, Vincent D, Bacot F, Ambrosone CB, Bandera EV, John EM, Chen GK, Hu JJ, Rodriguez-Gil JL, Bernstein L, Press MF, Ziegler RG, Millikan RM, Deming-Halverson SL, Nyante S, Ingles SA, Waisfisz Q, Tsimiklis H, Makalic E, Schmidt D, Bui M, Gibson L, Muller-Myhsok B, Schmutzler RK, Hein R, Dahmen N, Beckmann L, Aaltonen K, Czene K, Irwanto A, Liu J, Turnbull C, Familial Breast Cancer S, Rahman N, Meijers-Heijboer H, Uitterlinden AG, Rivadeneira F, Australian Breast Cancer Tissue Bank I, Olswold C, Slager S, Pilarski R, Ademuyiwa F, Konstantopoulou I, Martin NG, Montgomery GW, Slamon DJ, Rauh C, Lux MP, Jud SM, Bruning T, Weaver J, Sharma P, Pathak H, Tapper W, Gerty S, Durcan L, Trichopoulos D, Tumino R, Peeters PH, Kaaks R, Campa D, Canzian F, Weiderpass E, Johansson M, Khaw KT, Travis R, Clavel-Chapelon F, Kolonel LN, Chen C, Beck A, Hankinson SE, Berg CD, Hoover RN, Lissowska J, Figueroa JD, Chasman DI, Gaudet MM, Diver WR, Willett WC, Hunter DJ, Simard J, Benitez J, Dunning AM, Sherman ME, Chenevix-Trench G, Chanock SJ, Hall P, Pharoah PD, Vachon C, Easton DF, Haiman CA, Kraft P (2013) Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet 45(4):392–398, 398e391-392. https://doi.org/10.1038/ng.2561
Zhou L, Zhang X, Li Z, Zhou C, Li M, Tang X, Lu C, Li H, Yuan Q, Yang M (2013) Association of a genetic variation in a miR-191 binding site in MDM4 with risk of esophageal squamous cell carcinoma. PLoS One 8(5):e64331. https://doi.org/10.1371/journal.pone.0064331
Xu C, Zhu J, Fu W, Liang Z, Song S, Zhao Y, Lyu L, Zhang A, He J, Duan P (2016) MDM4 rs4245739 A > C polymorphism correlates with reduced overall cancer risk in a meta-analysis of 69477 subjects. Oncotarget 7(44):71718–71726. https://doi.org/10.18632/oncotarget.12326
Gansmo LB, Romundstad P, Birkeland E, Hveem K, Vatten L, Knappskog S, Lonning PE (2015) MDM4 SNP34091 (rs4245739) and its effect on breast-, colon-, lung-, and prostate cancer risk. Cancer Med 4(12):1901–1907. https://doi.org/10.1002/cam4.555
Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, van der Kwast T, Mason M, Matveev V, Wiegel T, Zattoni F, Mottet N, European Association of U (2014) EAU guidelines on prostate cancer. part 1: Screening, diagnosis, and local treatment with curative intent-update 2013. Eur Urol 65 (1):124–137. doi:https://doi.org/10.1016/j.eururo.2013.09.046
Sole X, Guino E, Valls J, Iniesta R, Moreno V (2006) SNPStats: a web tool for the analysis of association studies. Bioinformatics 22(15):1928–1929. https://doi.org/10.1093/bioinformatics/btl268
Preskill C, Weidhaas JB (2013) SNPs in microRNA binding sites as prognostic and predictive cancer biomarkers. Crit Rev Oncog 18(4):327–340. https://doi.org/10.1615/critrevoncog.2013007254
Moszynska A, Gebert M, Collawn JF, Bartoszewski R (2017) SNPs in microRNA target sites and their potential role in human disease. Open Biol 7(4):170019. https://doi.org/10.1098/rsob.170019
Pelletier C, Weidhaas JB (2010) MicroRNA binding site polymorphisms as biomarkers of cancer risk. Expert Rev Mol Diagn 10(6):817–829. https://doi.org/10.1586/erm.10.59
Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin Y, Wang E, Wu M, Shen SH (2007) Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res 35(13):4535–4541. https://doi.org/10.1093/nar/gkm480
Ding WH, Ren KW, Yue C, Zou JG, Zuo L, Zhang LF, Bai Y, Okada A, Yasui T, Mi YY (2018) Association between three genetic variants in kallikrein 3 and prostate cancer risk. Biosci Rep 38(6). https://doi.org/10.1042/BSR20181151
Zong H, Wang CC, Vaitheesvaran B, Kurland IJ, Hong W, Pessin JE (2011) Enhanced energy expenditure, glucose utilization, and insulin sensitivity in VAMP8 null mice. Diabetes 60(1):30–38. https://doi.org/10.2337/db10-0231
Kote-Jarai Z, Olama AA, Giles GG, Severi G, Schleutker J, Weischer M, Campa D, Riboli E, Key T, Gronberg H, Hunter DJ, Kraft P, Thun MJ, Ingles S, Chanock S, Albanes D, Hayes RB, Neal DE, Hamdy FC, Donovan JL, Pharoah P, Schumacher F, Henderson BE, Stanford JL, Ostrander EA, Sorensen KD, Dork T, Andriole G, Dickinson JL, Cybulski C, Lubinski J, Spurdle A, Clements JA, Chambers S, Aitken J, Gardiner RA, Thibodeau SN, Schaid D, John EM, Maier C, Vogel W, Cooney KA, Park JY, Cannon-Albright L, Brenner H, Habuchi T, Zhang HW, Lu YJ, Kaneva R, Muir K, Benlloch S, Leongamornlert DA, Saunders EJ, Tymrakiewicz M, Mahmud N, Guy M, O'Brien LT, Wilkinson RA, Hall AL, Sawyer EJ, Dadaev T, Morrison J, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Lophatonanon A, Southey MC, Hopper JL, English DR, Wahlfors T, Tammela TL, Klarskov P, Nordestgaard BG, Roder MA, Tybjaerg-Hansen A, Bojesen SE, Travis R, Canzian F, Kaaks R, Wiklund F, Aly M, Lindstrom S, Diver WR, Gapstur S, Stern MC, Corral R, Virtamo J, Cox A, Haiman CA, Le Marchand L, Fitzgerald L, Kolb S, Kwon EM, Karyadi DM, Orntoft TF, Borre M, Meyer A, Serth J, Yeager M, Berndt SI, Marthick JR, Patterson B, Wokolorczyk D, Batra J, Lose F, McDonnell SK, Joshi AD, Shahabi A, Rinckleb AE, Ray A, Sellers TA, Lin HY, Stephenson RA, Farnham J, Muller H, Rothenbacher D, Tsuchiya N, Narita S, Cao GW, Slavov C, Mitev V, Easton DF, Eeles RA, Oncology UKGPCSCBAoUSSo, Uk ProtecT Study Collaborators TAPCB, Consortium P (2011) Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet 43(8):785–791. https://doi.org/10.1038/ng.882
Li Q, Lozano G (2013) Molecular pathways: targeting Mdm2 and Mdm4 in cancer therapy. Clin Cancer Res 19(1):34–41. https://doi.org/10.1158/1078-0432.CCR-12-0053
Karni-Schmidt O, Lokshin M, Prives C (2016) The roles of MDM2 and MDMX in Cancer. Annu Rev Pathol 11:617–644. https://doi.org/10.1146/annurev-pathol-012414-040349
Wynendaele J, Bohnke A, Leucci E, Nielsen SJ, Lambertz I, Hammer S, Sbrzesny N, Kubitza D, Wolf A, Gradhand E, Balschun K, Braicu I, Sehouli J, Darb-Esfahani S, Denkert C, Thomssen C, Hauptmann S, Lund A, Marine JC, Bartel F (2010) An illegitimate microRNA target site within the 3' UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res 70(23):9641–9649. https://doi.org/10.1158/0008-5472.CAN-10-0527
Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S, Dadaev T, Neal DE, Hamdy FC, Donovan JL, Muir K, Giles GG, Severi G, Wiklund F, Gronberg H, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Lindstrom S, Kraft P, Hunter DJ, Gapstur S, Chanock SJ, Berndt SI, Albanes D, Andriole G, Schleutker J, Weischer M, Canzian F, Riboli E, Key TJ, Travis RC, Campa D, Ingles SA, John EM, Hayes RB, Pharoah PD, Pashayan N, Khaw KT, Stanford JL, Ostrander EA, Signorello LB, Thibodeau SN, Schaid D, Maier C, Vogel W, Kibel AS, Cybulski C, Lubinski J, Cannon-Albright L, Brenner H, Park JY, Kaneva R, Batra J, Spurdle AB, Clements JA, Teixeira MR, Dicks E, Lee A, Dunning AM, Baynes C, Conroy D, Maranian MJ, Ahmed S, Govindasami K, Guy M, Wilkinson RA, Sawyer EJ, Morgan A, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As NJ, Woodhouse CJ, Thompson A, Dudderidge T, Ogden C, Cooper CS, Lophatananon A, Cox A, Southey MC, Hopper JL, English DR, Aly M, Adolfsson J, Xu J, Zheng SL, Yeager M, Kaaks R, Diver WR, Gaudet MM, Stern MC, Corral R, Joshi AD, Shahabi A, Wahlfors T, Tammela TL, Auvinen A, Virtamo J, Klarskov P, Nordestgaard BG, Roder MA, Nielsen SF, Bojesen SE, Siddiq A, Fitzgerald LM, Kolb S, Kwon EM, Karyadi DM, Blot WJ, Zheng W, Cai Q, McDonnell SK, Rinckleb AE, Drake B, Colditz G, Wokolorczyk D, Stephenson RA, Teerlink C, Muller H, Rothenbacher D, Sellers TA, Lin HY, Slavov C, Mitev V, Lose F, Srinivasan S, Maia S, Paulo P, Lange E, Cooney KA, Antoniou AC, Vincent D, Bacot F, Tessier DC, Initiative CO-CRUG-E, Australian Prostate Cancer B, Oncology UKGPCSCBAoUSSo, Collaborators UKPS, Consortium P, Kote-Jarai Z, Easton DF (2013) Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet 45(4):385–391, 391e381-382. https://doi.org/10.1038/ng.2560
Fan C, Wei J, Yuan C, Wang X, Jiang C, Zhou C, Yang M (2014) The functional TP53 rs1042522 and MDM4 rs4245739 genetic variants contribute to non-Hodgkin lymphoma risk. PLoS One 9(9):e107047. https://doi.org/10.1371/journal.pone.0107047
Hashemi M, Sanaei S, Hashemi SM, Eskandari E, Bahari G (2018) Association of Single Nucleotide Polymorphisms of the MDM4 Gene with the susceptibility to breast Cancer in a southeast Iranian population sample. Clin Breast Cancer 18(5):e883–e891. https://doi.org/10.1016/j.clbc.2018.01.003
Mohammad Khanlou Z, Pouladi N, Hosseinpour Feizi M, Pedram N (2017) Lack of associations of the MDM4 rs4245739 polymorphism with risk of thyroid Cancer among Iranian-Azeri patients: a case-control Study. Asian Pac J Cancer Prevent: APJCP 18(4):1133–1138. https://doi.org/10.22034/APJCP.2017.18.4.1133
Pedram N, Pouladi N, Feizi MA, Montazeri V, Sakhinia E, Estiar MA (2016) Analysis of the association between MDM4 rs4245739 single nucleotide polymorphism and breast Cancer susceptibility. Clin Lab 62(7):1303–1308. https://doi.org/10.7754/Clin.Lab.2016.151128
Zhai Y, Dai Z, He H, Gao F, Yang L, Dong Y, Lu J (2016) A PRISMA-compliant meta-analysis of MDM4 genetic variants and cancer susceptibility. Oncotarget 7(45):73935–73944. https://doi.org/10.18632/oncotarget.12558
Purrington KS, Slager S, Eccles D, Yannoukakos D, Fasching PA, Miron P, Carpenter J, Chang-Claude J, Martin NG, Montgomery GW, Kristensen V, Anton-Culver H, Goodfellow P, Tapper WJ, Rafiq S, Gerty SM, Durcan L, Konstantopoulou I, Fostira F, Vratimos A, Apostolou P, Konstanta I, Kotoula V, Lakis S, Dimopoulos MA, Skarlos D, Pectasides D, Fountzilas G, Beckmann MW, Hein A, Ruebner M, Ekici AB, Hartmann A, Schulz-Wendtland R, Renner SP, Janni W, Rack B, Scholz C, Neugebauer J, Andergassen U, Lux MP, Haeberle L, Clarke C, Pathmanathan N, Rudolph A, Flesch-Janys D, Nickels S, Olson JE, Ingle JN, Olswold C, Slettedahl S, Eckel-Passow JE, Anderson SK, Visscher DW, Cafourek VL, Sicotte H, Prodduturi N, Weiderpass E, Bernstein L, Ziogas A, Ivanovich J, Giles GG, Baglietto L, Southey M, Kosma VM, Fischer HP, Network G, Reed MW, Cross SS, Deming-Halverson S, Shrubsole M, Cai Q, Shu XO, Daly M, Weaver J, Ross E, Klemp J, Sharma P, Torres D, Rudiger T, Wolfing H, Ulmer HU, Forsti A, Khoury T, Kumar S, Pilarski R, Shapiro CL, Greco D, Heikkila P, Aittomaki K, Blomqvist C, Irwanto A, Liu J, Pankratz VS, Wang X, Severi G, Mannermaa A, Easton D, Hall P, Brauch H, Cox A, Zheng W, Godwin AK, Hamann U, Ambrosone C, Toland AE, Nevanlinna H, Vachon CM, Couch FJ (2014) Genome-wide association study identifies 25 known breast cancer susceptibility loci as risk factors for triple-negative breast cancer. Carcinogenesis 35(5):1012–1019. https://doi.org/10.1093/carcin/bgt404
Stevens KN, Vachon CM, Couch FJ (2013) Genetic susceptibility to triple-negative breast cancer. Cancer Res 73(7):2025–2030. https://doi.org/10.1158/0008-5472.CAN-12-1699
Liu J, Tang X, Li M, Lu C, Shi J, Zhou L, Yuan Q, Yang M (2013) Functional MDM4 rs4245739 genetic variant, alone and in combination with P53 Arg72Pro polymorphism, contributes to breast cancer susceptibility. Breast Cancer Res Treat 140(1):151–157. https://doi.org/10.1007/s10549-013-2615-x
Acknowledgments
The research was supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia (Project no. 173016).
Funding
The study was financially supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia (Contract no. 451-03-68/2020-14/ 200178).
Author information
Authors and Affiliations
Contributions
N.K. performed genetic analysis and wrote the manuscript. Z.D. performed the statistical analysis and reviewed the manuscript. S.M. supported genetic analysis. D.S.P. and GB reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that there is no conflict of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kotarac, N., Dobrijevic, Z., Matijasevic, S. et al. Association of KLK3, VAMP8 and MDM4 Genetic Variants within microRNA Binding Sites with Prostate Cancer: Evidence from Serbian Population. Pathol. Oncol. Res. 26, 2409–2423 (2020). https://doi.org/10.1007/s12253-020-00839-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-020-00839-7