Summary
The digitate-tubered clade (Dactylorhiza s.l. plus Gymnadenia s.l.) within subtribe Orchidinae is an important element of the North-temperate orchid flora and has become a model system for studying the genetic and epigenetic consequences of organism-wide ploidy change. Here, we integrate morphological phylogenetics with Sanger sequencing of nrITS and the plastid region trnL-F in order to explore phylogenetic relationships and phenotypic character evolution within the clade. The resulting morphological phylogenies are strongly incongruent with the molecular phylogenies, instead reconstructing through parsimony the genus-level boundaries recognised by traditional 20th Century taxonomy. They raise fresh doubts concerning whether Pseudorchis is sister to Platanthera or to Dactylorhiza plus Gymnadenia. Constraining the morphological matrix to the topology derived from ITS sequences increased tree length by 20%, adding considerably to the already exceptional level of phenotypic homoplasy. Both molecular and morphological trees agree that D. viridis and D. iberica are the earliest-diverging species within Dactylorhiza (emphasising the redundancy of the former genus Coeloglossum). Morphology and ITS both suggest that the former genus Nigritella is nested within (and thus part of) Gymnadenia, the Pyrenean endemic 'N.' gabasiana apparently forming a molecular bridge between the two radically contrasting core phenotypes. Comparatively short subtending molecular branches plus widespread (though sporadic) hybridisation indicate that Dactylorhiza and Gymnadenia approximate the minimum level of molecular divergence acceptable in sister genera. They share similar tuber morphologies and base chromosome numbers, and both genera are unusually prone to polyploid speciation. Another prominent feature of multiple speciation events within Gymnadenia is floral paedomorphosis. The 'traditional' morphological and candidate-gene approaches to phylogeny reconstruction are critically appraised.
Similar content being viewed by others
Introduction
Subtribe Orchidinae s.s. (i.e. excluding the still poorly resolved subtribe Habenariinae) dominates the Eurasian orchid flora, encompassing a considerable range of phenotypes (Fig. 1) and evolutionary mechanisms (reviewed by Bateman 2009, 2012a). This clade has, through the last two decades, been subjected to several molecular phylogenetic studies utilising many samples that together spanned the subtribe (e.g. Bateman et al. 2003; Inda et al. 2012; Jin et al. 2014; Tang et al. 2015; Jin et al. 2017). Several genera within the subtribe have also been subjected individually to more detailed molecular phylogenetic examination; these include Ophrys (Soliva & Widmer 2003; Devey et al. 2008; Breitkopf et al. 2015; Bateman et al. 2018), Serapias (Bellusci et al. 2008), Himantoglossum s.l. (Sramkó et al. 2014), Orchis s.s. (Tyteca et al. 2012), Platanthera (Hapeman & Inoue 1997; Bateman et al. 2009), Hemipilia s.l. (Jin et al. 2014; Tang et al. 2015), Dactylorhiza (Devos et al. 2006a; Pillon et al. 2007; Hedrén et al. 2011) and Gymnadenia (Bateman et al. 2006; Stark et al. 2011; Efimov 2013; Sun et al. 2015; Hedrén et al. 2018).
A Platanthera hyperborea (Iceland); B Galearis spectabilis (Maryland, U.S.A.); C Dactylorhiza incarnata subsp. pulchella (England); D D. viridis (England); E Gymnadenia (Nigritella) austriaca (left), G. conopsea s.s. (centre), G. odoratissima (right) (co-occurring in the Italian Dolomites); F D. romana (yellow-flowered morph, left), Orchis provincialis (right) (Sicily); G G. densiflora (U.K.); H G. borealis (England); J G. frivaldii (N Greece); K Pseudorchis albida (Scotland). photos: richard bateman.
This study is focused at an intermediate taxonomic level, seeking to clarify (a) genus-level relationships within the taper-tuber clade sensu Bateman et al. (2006) (i.e. those genera that do not rely entirely on roots emerging near-horizontally from the base of the stem, but also emit at least one large near-vertical root from the apex of the consequently tapered tuber: Platanthera, Galearis, Neolindleya, Pseudorchis, Dactylorhiza, Gymnadenia) and (b) species-level relationships (and the underlying causes of speciation) within Dactylorhiza s.l. (including the former genus Coeloglossum) and Gymnadenia s.l. (including the former genus Nigritella). These two genera have been consistently found to be sisters; they have been shown to share digitate tubers (i.e., those emitting more than one large root from the tuber apex) and a presumed chromosomal fusion event that converted n = 21 to n = 20 (Pridgeon et al. 1997; Bateman et al. 2003). These genera were segregated as subtribe Dactylorhizinae by Vermeulen (1977).
Trees based on Sanger sequencing of nuclear ribosomal ITS and the plastid region trnL-F are compared with trees generated via a morphological cladistic analysis spanning the subtribe, with the dual key objectives of (a) comparing these contrasting phylogenetic signals and (b) elucidating the sequence of acquisition of phenotypic characters within the taper-tubered clade in general and the digitate-tubered clade in particular. In addition to further exploring pattern and process within this particular clade of orchids, this study constitutes an introspective re-examination of the strengths and weaknesses evident in what has become a traditional — arguably even passé — approach to phylogeny reconstruction.
Materials and Methods
Fieldwork directed toward this study occurred sporadically across the northern hemisphere from 1996 onward (RB, ID, PR and several associates), collecting samples in the now ubiquitous silica gel sachets. Laboratory work took place in three separate phases: in the late 1990s at RBG Edinburgh (RB, PH, MH), in the early 2000s at NHM London (RB, K. James: Bateman et al. 2006), and in 2017 at RBG Kew (RB, PR, AM); some details of the earlier phases of laboratory analyses have consequently been lost. Samples that yielded novel DNA sequences generated specifically for the present project are summarised in Table 1.
Data generation: nuclear ITS
The complete nuclear ribosomal ITS region was amplified using primers modified from White et al. (1990): “ITS 5p” (5’–GGAAGGAGAAGTCGTAACAAG) and “ITS 4p” (5’–TCCTCCGCTTATTGATATGC). PCR reactions of 50 μl contained 2 μl DNA template, 100 μM of each dNTP, 0.3 μM of each primer, 2 units Taq polymerase (Bioline), 2 μM MgCl2 and 5 μl reaction buffer (160 mM (NH4)2SO4, 670 mM Tris HCl, 0.1% Tween 20, pH 8.8). The following PCR protocol was used: 1 cycle at 94°C for 3 minutes; 30 cycles at 94°C for 1 min, 55°C for 1 min and 72°C for 90 sec; 1 cycle at 72°C for 5 min. The resulting PCR products were purified using Qiagen PCR purification kits according to the manufacturer's instructions. Sequencing PCR was performed using Thermosequenase II TM dye terminator sequencing premix kit (Amersham Pharmacia, UK) according to the manufacturer’s recommendations. Sequencing reactions were run on an ABI 377 automated sequencer and the output files were edited using Sequence Navigator (Applied Biosystems Inc.).
Data generation: plastid trnL-F
DNA was extracted from single bracts using a protocol modified from Doyle & Doyle (1990). A ground-glass rod attached to a domestic power drill was used to homogenise samples in 2× CTAB buffer (pre-heated to 65°C), with 0.2 % β-mercaptoethanol along with a pinch of polyvinylpolypyrrolidone (PVPP) and acid-washed sand. Samples were incubated at 65°C for 60 min. An equal volume of 24: 1 chloroform/isoamyl alcohol was then added and the samples were spun for 10 min at 13,000 rpm in a microfuge. The supernatant was removed and the chloroform/isoamyl alcohol step repeated. Following centrifugation, the supernatant was decanted, and the DNA precipitated by the addition of two-thirds volume freezer-cold isopropanol. The samples were then centrifuged at 13,000 rpm for 10 min to collect the resulting pellet. Finally, the isopropanol was decanted, and after air-drying for 20 min, the DNA was re-suspended in 300 μL of tris-ethylenediaminetetraacetic acid (TE) prior to sequencing, which encompassed the trnL intron, trnL-F intergenic spacer and the short intervening exon.
Data generation: Morphology
The morphological cladistic matrix was compiled from a wide range of sources with the primary aim of describing all aspects of the plants' phenotype that could realistically be summarised as qualitative (bistate or multistate) rather than quantitative (metric or meristic) characters. When selecting characters, no prior consideration was given to the likelihood of encountering extensive homoplasy, although character selection did attempt to minimise the risk of character duplication caused by underlying pleiotropy. The range of coded taxa was selected to broadly correspond with that — despite the near-typological sampling — represented in the corresponding ITS matrix. However, once it became evident that the outgroup genera selected initially (two species of Platanthera s.l., one each of Galearis and Neolindleya) did not resolve as monophyletic in the morphological tree, we then scored two additional, more phylogenetically distant outgroups (Orchis, Hemipilia s.l.).
Character states were obtained from the extensive literature (Vermeulen 1972; Luer 1975; Landwehr 1977; Strack et al. 1989; Luo & Chen 2000; Barone-Lumaga et al. 2006; Bateman et al. 2006; Delforge 2006; Box et al. 2008; Gamarra et al. 2008; Bateman et al. 2009; Bell et al. 2009; Chen et al. 2009; Efimov et al. 2009; Claessens & Kleynen 2011; Bateman et al. 2015; Gamarra et al. 2015; Tang et al. 2015; Bateman & Rudall 2018), liberally supplemented with our own observations based on the study of living plants, dried specimens held in the RBG Kew herbarium, and scanning electron microscopy performed in Kew's Jodrell Laboratory (Fig. 2).
Scanning electron micrographs of representative Orchidinae flowers (most are open flowers, although H and J are dissected flower buds). A Platanthera bifolia, gynostemium and proximal portion of labellum, featuring spur opening and lateral auricles; B P. bifolia, inside of spur, showing elongate papillae; C Dactylorhiza iberica, gynostemium, showing prominent bursicle, vertical rostellum and expansive thecae; D D. viridis, gynostemium and proximal portion of labellum, showing spur opening, suprajacent stigma, broad gynostemium and paired viscidia; E Gymnadenia densiflora, gynostemium and proximal portion of labellum, showing spur opening and lateral stigmatic surfaces encrusted in pollinium massulae; F Pseudorchis albida, showing compact gynostemium; G Ps. straminea, flower illustrating petals and sepals; H Gymnadenia (Nigritella) austriaca, immature compact gynostemium and longitudinally sectioned spur lacking papillae; J G. densiflora, immature gynostemium with labellum removed to reveal developing elongate spur; K G. conopsea, flower illustrating petals and sepals; L G. frivaldii, flower illustrating petals (including comparatively undifferentiated labellum) and sepals, paedomorphic relative to K; Labels: au auricle; bu bursicle; ca caudicle; co connective; ds dorsal sepal; gy gynostemium; la labellum; ls lateral sepal; lp lateral petal; ov ovary; po pollinium; ro rostellum; sc cordate stigma; spe spur entrance; stl lateral stigma lobe (bilobed stigma 'lappets'); sp labellar spur; th theca; vi viscidium. Scale bars: A, D, G = 1 mm; C, E, I = 250 μm; B, F, H, J, K = 100 μm. micrographs: paula rudall.
Preparation for SEM involved selecting flowers from each preserved inflorescence for dehydration through an alcohol series to 100% ethanol. They were then stabilised using an Autosamdri 815B critical-point drier, mounted onto stubs using double-sided adhesive tape, coated with platinum using an Emtech K550X sputter-coater, and examined under a Hitachi cold-field emission SEM S-4700-II at 2 kV. The resulting images were recorded digitally for subsequent enhancement in Adobe Photoshop.
After considerable experimentation, 51 morphological characters were scored for 27 coded taxa, thereby generating a matrix of 1,377 cells. The characters (listed in Table 2) encompassed cytology, breeding system, and the morphology of every organ of the plant, the 13 vegetative characters being numerically subordinate to 35 floral characters. A total of 47 cells (3.4%), distributed among 14 of the 51 characters, were coded as polymorphic (i.e. containing two or more character states). The most severely affected characters were labellum colour (C36: 10 of the 27 cells), leaf number (C10: 7 cells) and leaf arrangement along the stem (C13: 7 cells). No organ was represented by fewer than two characters. A further 133 cells (9.6%) were coded as missing, including a few cells in which the character was inapplicable for that species. In this case, the most severely affected characters were all micromorphological: spur papilla shape (C26: 18 cells), pollen exine surface sculpture (C51: 16 cells), spur interior striations (C24: 14 cells) and labellum adaxial epidermal cell morphology (C38: 14 cells). These figures are significant because the influence on tree building of any character containing more than approximately one-third polymorphic or missing values is considerably reduced.
Tree-building: ITS
Data for both the ITS and trnL-F were entered as Fasta files into MacClade v4.05 (Maddison & Maddison 2002), aligned by eye, and each of the respective resulting matrices was subjected to parsimony analyses in PAUP 4.0b10 (Swofford 2003), supported by GTR maximum likelihood (ML) analyses. Parsimony analyses were conducted as heuristic searches under TBR branch swapping and the amb– collapse criterion. Subsequent bootstrap analyses involved 1,000 replicates for trnL-F and 500 replicates for ITS for parsimony analyses, but was reduced to 100 replicates for both matrices in the more time-consuming ML analyses. Gaps were treated as missing, the less ambiguous among the apparent indels being coded as independent characters largely following the "simple coding" logic of Simmons & Ochoterena (2000).
Our initial ITS matrix combined 83 sequences generated for the present study or its direct predecessors (Pridgeon et al. 1997; Bateman et al. 2003; Bateman et al. 2006; Pillon et al. 2006; Pillon et al. 2007; Bateman et al. 2015; Tang et al. 2015) with a smaller number of sequences downloaded from studies published by several other research groups (Soliva & Widmer 1999; Stark et al. 2011; Jin et al. 2012; Efimov 2013; Sonkoly et al. 2016). The accumulated matrix of 129 sequences was then pared down to 46 representative ribotypes that exhibited non-polymorphic differences in nucleotides or indels, plus one sequence from Gotland showing several polymorphisms that clearly demonstrated the presence of two ribotypes that presumably reflect recent hybridisation between two Gymnadenia species. (Note: we are aware that the term "ribotype" was originally coined to describe the less complex ITS sequences of prokaryotes; nonetheless, we find it a useful shorthand across all the biotic kingdoms for "nrITS sequence variant".)
Tree-building: trnL-F
Most of the 45 trnL-F sequences initially collated were generated specifically for the present study at Royal Botanic Garden Edinburgh, although eight were downloaded from GenBank (between one and four sequences each derived from the studies of Soliva & Widmer 1999; Soininen et al. 2009; Tang et al. 2015; Pornon et al. 2016). The overall alignment of the matrix required 712 base positions plus a further 20 indels. Two sequences lacked the initial 168 bp (Chinese samples of Gymnadenia orchidis and Dactylorhiza viridis) and one sequence lacked the terminal 138 bp (Pseudorchis straminea). One phylogenetically important species present in the ITS matrix, G. gabasiana, was absent from the trnL-F matrix, wherein the many controversial allotetraploid Dactylorhiza species were represented as a placeholder by D. cordigera rather than D. majalis. The 45 available sequences were pared down to 21 core sequences that differed in SNPs and/or indels; these 21 haplotypes formed the basis of subsequent tree-building, after setting aside a poly-A region located near the 5' end of the trnL intron (first reported by Soliva & Widmer 1999) that varied in length from 9 bp to 18 bp and failed to correlate with any credible taxonomy.
Tree building: morphology
The data presented as Table 3 — including polymorphic cells — were entered into MacClade v4.05 (Maddison & Maddison 2002) and analysed in PAUP 4.0b10 (Swofford 2003) using both parsimony and the widely used (if frequently criticised) phenetic algorithms neighbour joining (NJ) and unweighted pair group method with arithmetic mean (UPGMA). Parsimony analyses were conducted as heuristic searches under the amb– collapse criterion and Acctran optimisation. The subsequent bootstrap analysis involved 1,000 replicates.
Results
TrnL-F phylogeny
Of the 732 positions plus indels analysed for 21 haplotypes, only 59 were variable and only 22 (including 11 indels) were parsimony informative. The resulting single most-parsimonious tree (shown under Acctran optimisation as Fig. 3) was 74 steps long, yielding a consistency index of 0.816 (0.611 excluding uninformative characters) and a retention index of 0.823. The node separating Gymnadenia orchidis from the remaining Gymnadenia species reflected only a single homoplastic SNP and hence collapsed in the strict consensus tree (it was also polytomous in the equivalent likelihood tree: not shown). The three ingroup genera all received at least 68% bootstrap support in the parsimony tree, but the node that identified Dactylorhiza and Gymnadenia s.l. as sister genera relative to Pseudorchis received only 50% (and less than 50% in the equivalent ML tree), reflecting the presence of just one non-homoplastic indel. The greatest separation was evident between the outgroup Platanthera and the ingroup, the observed disparity consisting of eight SNPs plus four indels.
Single most-parsimonious tree of the taper-tubered clade of subtribe Orchidinae, generated via maximum parsimony from the trnL-F (plastid) matrix. Acctran optimisation. Asterisk indicates the node that collapsed in the strict (and also majority rule) consensus tree. The six branches that achieved bootstrap support values exceeding 50% are indicated in roman script; equivalent bootstrap values obtained through ML analysis are italicised. Haplotypes within Gymnadenia s.l. are distinguished by roman numerals.
Much of the phylogenetic signal in trnL-F was provided by a 9 bp minisatellite region (typical motif ATAATAGTA) located midway through trnL-F; this occurred once in Dactylorhiza, Pseudorchis and (in a modified motif) in the Platanthera outgroup, twice or thrice in 'classic' Gymnadenia, but was absent from the former genus Nigritella. The apparent deletion event of this minisatellite from the former genus Nigritella and the apparent insertion event in the remainder of Gymnadenia were the only characters separating the two groups, thereby inevitably leading to their poorly-supported representation as sister groups (Fig. 3).
Little variation (and thus little resolution) was evident within any of the genera analysed. All members of the former genus Nigritella yielded identical sequences once the poly-A region had been excluded. Gymnadenia orchidis deviated from the remainder primarily in four indels, although it also possessed a "rogue" SNP that otherwise characterised both Dactylorhiza and 'Nigritella'. Remarkably, a single haplotype characterised the majority of individuals analysed of each of the other species of 'core' Gymnadenia analysed (60% of conopsea, 63% of odoratissima, 100% of borealis, 60% of densiflora). Other haplotypes observed in few individuals of each species deviated from the core haplotype by only one or two autapomorphies (the majority of them indels: Fig. 3).
ITS phylogeny
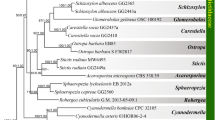
Of the 660 positions plus 12 indels analysed for 47 ribotypes, 166 proved variable and 117 were parsimony informative (including seven indels). The resulting 179 most-parsimonious trees of length 251 yielded a consistency index of 0.783 (0.729 excluding uninformative characters) and a retention index of 0.908. A representative tree is shown as Fig. 4. Of the three nodes that collapsed in the 50% majority rule consensus tree, two had only trivial implications: that supporting the pairings of Dactylorhiza fuchsii plus D. majalis and that supporting the rare ribotypes VII and VIII of Gymnadenia conopsea. However, the third ambiguous node was of greater significance: it was equally parsimonious to place G. frivaldii as sister to G. densiflora (as in Fig. 4) or as sister to G. borealis. The equivalent likelihood tree (not shown) differed only in collapsing the node shared by the two species of Pseudorchis in the parsimony tree.
Representative example of 179 maximum parsimony trees of the taper-tubered clade of subtribe Orchidinae, generated via maximum parsimony from the ITS (nuclear ribosomal) matrix. Acctran optimisation. Asterisks indicate nodes that collapsed in the strict (and also majority rule) consensus tree. The 26 branches that achieved bootstrap support values exceeding 50% are indicated in roman script; equivalent bootstrap values obtained through ML analysis are italicised. Ribotypes within Gymnadenia s.l. are distinguished by roman numerals (core ribotypes are emphasised in boldface), and the geographic origins of the relevant samples are also shown.
Few branches attracted more than 50% bootstrap support under parsimony criteria, and even fewer under likelihood. Platanthera was clearly an appropriate outgroup, and the monophyly of each of the three ingroup genera is supported with at least 71% bootstrap support. Pseudorchis is placed as sister to the two remaining ingroup genera with 100% support (Fig. 4). Sampling of Dactylorhiza was largely typological, only the especially phylogenetically controversial D. viridis being represented by multiple accessions. In contrast, comparatively intensive sampling of Gymnadenia s.l. identified a total of 30 ribotypes, representing 13 putative species. The six analysed species of the former genus Nigritella collectively yielded only two ribotypes, whereas nine putative ribotypes were found within G. densiflora alone and eight within G. conopsea s.s. Remarkably, three ribotypes were shared between the phenotypically distinct species G. conopsea and G. odoratissima.
Morphological phylogenies
Of the 51 coded characters analysed for 27 representative species of Orchidinae, all 51 were variable but only 46 were parsimony informative. The resulting 10 most-parsimonious trees of length 181 yielded a consistency index of 0.475 (0.457 excluding uninformative characters) and a retention index of 0.666. A representative tree is shown as Fig. 5. Only two comparatively unimportant nodes collapsed in the strict consensus tree: that linking Dactylorhiza foliosa with D. fuchsii, and that supporting a clade composed of D. incarnata, D. euxina, D. aristata and D. majalis (the allotetraploid derivative of D. fuchsii and D. incarnata). Despite the small number of most-parsimonious trees, the topological structure of the trees proved exceptionally fragile; only three nodes gained bootstrap values in excess of 50%, and only that underpinning the former genus Nigritella exceeded 75% (Fig. 5). The explanation for this topological weakness lies in the exceptionally low values for the retention index and especially the consistency index; only two individual character-state transitions were both non-homoplastic and non-autapomorphic (i.e. unambiguous synapomorphies): the absence of resupination throughout the former genus Nigritella (C18) and the shared presence of a central ridge on the labella of D. iberica and D. viridis (C35).
Representative example of the 10 maximum parsimony trees of the taper-tubered clade of subtribe Orchidinae, generated from the morphological matrix given in Table 2. Acctran optimisation. Asterisks indicate nodes that collapsed in the strict (and also majority rule) consensus trees. The three branches that achieved bootstrap support values exceeding 50% are indicated.
Neither the pair of Platanthera species nor the putative pairing of Galearis and Neolindleya species formed monophyletic groups (Fig. 5). Next to diverge was the pair of primitive Dactylorhiza species, D. iberica and D. viridis. The more derived species of Dactylorhiza emerged as sister to Gymnadenia s.l. (including the former genus Nigritella). However, Orchis mascula was embedded within Dactylorhiza as sister to D. sambucina, and Ps. albida was embedded within Gymnadenia as sister to G. frivaldii. Within the more derived Dactylorhiza species, D. fuchsii and D. foliosa were separated from the remainder. Within Gymnadenia, G. orchidis was identified as sister to the deeply embedded former genus Nigritella.
Phenetic trees were also generated from the morphological matrix in PAUP using the NJ (Fig. 6A) and UPGMA (Fig. 6B) algorithms. Both deviated substantially from the most-parsimonious topologies. In the NJ tree, Galearis spectabilis was promoted to a position as sister to Dactylorhiza iberica, the pairing of Platanthera hyperborea and Neolindleya camtschatica diverged later than the core Dactylorhiza clade, Pseudorchis was placed as sister to Gymnadenia, and G. frivaldii replaced G. orchidis as sister to the former genus Nigritella. The UPGMA tree diverged most radically from the parsimony trees, yielding a highly improbable topology. Most notably, the former genus Nigritella diverged earliest, reflecting the large number of character-state transitions that occur on its subtending branch. The pairing of Galearis spectabilis and D. iberica was next to diverge, followed by D. viridis as an isolated lineage. Orchis mascula was shown as sister to the remainder of Dactylorhiza, and Pseudorchis was shown as sister to the non-nigritellan portion of Gymnadenia s.l.
Having obtained these diverse, weakly supported topologies, we decided to map the morphological cladistic matrix across the topology recovered from the ITS matrix (Fig. 7). In order to generate the required backbone constraint prior to analysis in PAUP we combined four pairs of morphologically very similar species present in the ITS tree (D. saccifera into D. fuchsii, D. romana into D. sambucina, D. cf. hatagirea into D. incarnata, G. gabasiana into G. rhellicani), and added as more distant outgroups Galearis, the monotypic genus Neolindleya, Orchis s.s. and Hemipilia s.l. (the latter used in the original morphological cladistic analysis). Constraining the morphological characters to the molecular topology added 20% to the length of the most parsimonious tree(s), the increase to 218 steps further reducing the ensemble consistency index to 0.395 (0.377), the ensemble retention index to 0.535, and the total number of non-homoplastic synapomorphies to just four.
Morphological character-state transitions (Acctran optimisation) dictated by constraining the morphological cladistic matrix to the preferred topology generated from the ITS matrix (presented here as Fig. 4) The initial number matches characters listed in Table 2, whereas the following numbers indicate the transition between ancestral and derived states of that character. Italicised characters are homoplastic, boldface characters are non-homoplastic.
Consistency indices from the constraint tree were tabulated for each of the 51 morphological characters (Table 4) and mean values were calculated for each organ of the plant (including the five characters either fixed or optimised as autapomorphic). Mean consistency indices were also calculated for the three pigmentation values (C16, 36, 37: mean = 0.592) and the seven cellular-level micromorphological characters (C20, 24 – 26, 38, 50, 51: mean = 0.514). Homoplasy proved to be lowest in the character sets representing cytology and rootstocks, and greatest in those representing stem plus inflorescence, fragrance and nectar, labellar spurs, lateral petal/sepal positions, gynostemium morphology and (once adjusted for an unusually large number of ambiguous cells) seed/pollen morphology. Specific characters yielding consistency index values of less than 0.3 were breeding system (C3), stem architecture (C8), leaf arrangement (C12, C13), pedicel length (C17), spur papilla size (C25), spur length and diameter (C27, C28), pollinarium caudicle length (C44) and prominence of rostellar median fold (C48).
Discussion
Circumscription of, and relationships among, genera
Summary of previous molecular phylogenetic analyses — Fig. 8 compares topologies from four previous molecular phylogenetic studies of subtribe Orchidinae. Given the substantial proportion of branches lacking strong statistical support and thus incurring considerable uncertainty, it is perhaps unsurprising that considerable topological incongruence is evident between the various studies. Only two nodes persist across all five trees, but fortunately, they are the two nodes of greatest relevance to the present study: that linking Dactylorhiza and Gymnadenia as sister genera (usually with strong bootstrap support), and that linking this sister pairing to the remainder of the taper-tubered clade (i.e. Platanthera plus Galearis plus Neolindleya plus Pseudorchis). All five trees also agree, with strong bootstrap support, that both Dactylorhiza s.l. and Gymnadenia s.l. are monophyletic. Four of the trees agree that Pseudorchis is sister to Platanthera plus Galearis plus Neolindleya, and although the fifth (Tang et al. 2015) places Pseudorchis as sister to Platanthera, this inferred relationship lacks bootstrap support.
Topological comparison of five previous molecular phylogenetic analyses of subtribe Orchidinae s.s. (A – E). All trees were generated using Bayesian inference except that of Bateman et al. (maximum parsimony). Nuclear regions are shown in roman font, plastid regions are italicised, and mitochondrial regions are underlined. Dashed lines were represented only by a single accession. Statistical support is uniformly given as bootstrap values calculated via maximum parsimony; that supporting the taper-tubered clade is given in red font and that supporting the digitate-tubered clade in blue font. 'Traun+Cham' is Traunsteinera plus Chamorchis, the SE Asia clade is Hemipilia (including the former genera Neottianthe, Amitostigma, Ponerorchis and Chusua: Tang et al. 2015), and the 2n = 36 clade consists of Steveniella, Himantoglossum s.l., Anacamptis s.l., Serapias and Ophrys (Bateman et al. 2003). Note that the lower half of the topology of Jin et al. (2017) provides substantially different placements for Traunsteinera-Chamorchis (T), Neotinea (N) and the 2n = 36 clade (2n); also, the upper half of their topology nests 'Neolindleya' within Galearis s.s.
The apparent identity of the sister-group to the digitate-tubered clade differs greatly among trees, from Traunsteinera plus Chamorchis alone (Bateman et al. 2003) and Neotinea alone (Jin et al. 2017) through to a clade consisting of all of the remaining genera of Orchidinae other than Hemipilia s.l. (Inda et al. 2012). The morphologically mismatched Alpine pairing of Traunsteinera plus Chamorchis appears to be a particularly strong source of topological instability in these trees, as is Neolindleya within the Platanthera–Galearis clade (BS reliably <50%). Also noteworthy is the especially radical topological contrast between the two studies richest in "informative" bases: Inda et al. (2012) and Jin et al. (2017). Evidently, size is not everything.
Present molecular phylogenetic analysis — With the exception of the use of Orchis and Hemipilia as "outer outgroups" in the morphological cladistic analysis, the present study is confined to the six genera that constitute the taper-tubered clade (i.e. Dactylorhiza through to Galearis in Fig. 8). This narrower genus-level sampling inevitably weakens any conclusions regarding relationships between the genera. Nonetheless, we note that our attempts to use Pseudorchis as an outgroup alongside Platanthera reliably failed. In both the nuclear (Fig. 4) and plastid (Fig. 3) trees, Pseudorchis is placed as sister to Dactylorhiza plus Gymnadenia, contradicting its placement in every previous molecular phylogenetic tree as sister to Platanthera or, more commonly, sister to Platanthera plus Galearis/Neolindleya (Fig. 8). Moreover, the branch separating Pseudorchis from Dactylorhiza plus Gymnadenia is significantly shorter than that separating Pseudorchis from Platanthera in both of our molecular trees. In the morphological tree (Fig. 5), Pseudorchis is placed in an even more derived position, as sister to the basalmost unequivocal Gymnadenia species. And when the morphological data were constrained to a typical molecular topology (Fig. 7), Pseudorchis fitted poorly, being linked with Platanthera plus Galearis by only two homoplastic characters (facultatively autogamous breeding system, C3; white flowers, C36) that are inadequate to circumscribe the putative aggregate clade.
Another variable placement is that of the former genus Nigritella, which is nested deeply within Gymnadenia s.s. in the nuclear and morphology trees but placed as sister to Gymnadenia s.s. in the plastid tree — a distinction based entirely on the basis of contrasting, group-specific copy numbers of a distinctive 9 bp minisatellite. A corresponding sister-group relationship between these genera was derived through UPGMA analysis of allozyme data by Stahlberg (1999) and Hedrén et al. (2000), and through a median-joining network generated from haplotypes based on five plastid regions by Hedrén et al. (2018). The most likely scenario for our data suggests that the minisatellite was lost from 'Nigritella' but duplicated (and, in some cases, triplicated) in Gymnadenia s.s. On balance, the evidence remains strong for nesting of a monophyletic 'Nigritella' within a paraphyletic Gymnadenia s.s.
With regard to other former genera, both the present nuclear and plastid trees place the former genus Coeloglossum firmly within Dactylorhiza. It occupies this position in all of the previous studies summarised in Fig. 8; thus far, only Devos et al. (2006b) have ever succeeded in placing D. viridis (marginally) below the remainder of the genus Dactylorhiza in a molecular phylogeny (reviewed by Bateman & Rudall 2018). The morphological phylogenetic tree does place D. viridis below the remainder of Dactylorhiza plus Gymnadenia plus Pseudorchis, but transferred alongside D. viridis as putative sister is D. iberica. This species was never challenged by previous authors as being anything other than a fully-fledged member of the genus Dactylorhiza, and we would strongly discourage any future attempt to establish it as a monotypic genus (Bateman & Rudall 2018).
'Pseudorchis' frivaldii was omitted from the plastid tree in the absence of relevant sequences, although a previous study utilising multiple plastid sequences (Bateman et al. 2006) placed this species firmly within Gymnadenia, as does the present ITS tree. In contrast, the morphology tree places G. frivaldii as sister to Pseudorchis albida, thereby recovering its traditional genus-level assignment.
Taken at face value, the molecular phylogenies generated during the present study (Figs 3, 4) merely add to the complexity and ambiguity of relationships suggested by the previous studies here summarised in Fig. 8. Moreover, the present morphological cladogram — the first to be generated for the taper-tubered clade (Fig. 5) — deviates strongly from any of the relevant molecular topologies. Given these strongly contrasting topologies, the only overall conclusion permitted by pragmatism is that, although molecular phylogenetics has undoubtedly proven effective for re-circumscribing genera within Orchidinae, it has been rather less successful at determining with confidence the relationships between those re-circumscribed genera. Admittedly, morphology performs even less well than candidate-gene molecular studies.
Morphological cladistic analysis — Interestingly, in the case of Orchidinae, morphological cladistics has achieved neither credibly circumscribed genera nor clarified relationships. Nonetheless, the results of this experiment are of considerable interest. Firstly, our study illustrates the great importance to phylogeny reconstruction of thorough taxon sampling. Multiple molecular phylogenetic studies (Hapeman & Inoue 1997; Bateman et al. 2003; Bateman et al. 2009; Jin et al. 2014; Tang et al. 2015) have all made convincing arguments that Platanthera s.l. is monophyletic, yet in the present morphological study the two species of Platanthera selected as outgroups (P. chlorantha from section Platanthera, P. hyperborea from section Limnorchis) are sufficiently morphologically divergent to be separated phylogenetically by interpolation of Galearis spectabilis (Fig. 5). Moreover, Neolindleya — recently assigned to Galearis on molecular phylogenetic evidence by Jin et al. (2014) — is here shown as sister to P. hyperborea, the two species being linked by four ambiguously optimised but homoplastic characters: autogamous breeding system, stem central cavity, leaves well-distributed along stem, and loss of spur interior striations.
Within the ingroup, with one notable exception, the morphological cladogram (Fig. 5) reconstructs the superseded genus-level circumscriptions that dominated taxonomy of the group immediately prior to early applications of molecular phylogenetics (Pridgeon et al. 1997, et seq.). The former genus Coeloglossum is separated from the majority of Dactylorhiza species, albeit carrying with it D. iberica on the basis of reduced leaf number, shortened pedicel, labellum median ridge (non-homoplastic), lateral sepal connate with median sepal, and subdued median fold of rostellum. More radically, the putative outgroup Orchis mascula is instead placed within the ingroup as sister to D. sambucina, supported by two characters: expanded leaves concentrated into a basal rosette, and upwardly curved spur. Comparison of the flowers of co-occurring plants of D. romana (a close relative of D. sambucina) and O. provincialis (a close relative of O. mascula) shows remarkable evidence of convergence in several floral features (Fig. 1F).
Above Dactylorhiza, the morphological tree places Pseudorchis albida as sister to Gymnadenia, but detaches G. frivaldii from Gymnadenia and instead supports its more traditional placement within Pseudorchis, even though this placement is conclusively rejected by molecular studies showing their floral similarities to reflect convergence (Bateman et al. 2006). Characters supporting the pairing are reduced leaf number, shortened pedicel, greater concavity of the labellum, loss of doming in adaxial labellar epidermal cells. The one issue where the morphological cladogram differs from immediately pre-molecular circumscription is the placement of the former genus Nigritella; as previously shown in a preliminary morphological cladistic analysis (Bateman & DiMichele 2002), it is embedded deeply within Gymnadenia s.s. despite its subtending long branch. Here, it is linked to G. orchidis as sister by only one ambiguously optimised character state: short (i.e. reduced) caudicle length.
Given an initial assumption that traditional taxonomists are more likely to use overall morphological similarity than parsimony as a cerebral classificatory tool, we predicted from first principles that the pre-molecular genus circumscriptions would more likely emerge from the phenetic tree-building techniques NJ and/or UPGMA, but this proved to be a false assumption (cf. Figs 5, 6). Instead, the NJ tree pushed Dactylorhiza viridis and D. iberica even further from the remainder of Dactylorhiza and elevated Gymnadenia frivaldii further up the tree, to a position deeply embedded within Gymnadenia s.s. as sister to 'Nigritella' (Fig. 6A). In contrast, the UPGMA algorithm was seduced by the exceptionally long morphological branch subtending 'Nigritella' into placing this former genus in a highly improbable position as the earliest divergent among all of the taxa analysed (Fig. 6B). (Note: We are aware of the philosophical impurity inherent in the phrase 'early divergent', but nonetheless find it a useful shorthand for identifying within a topology particular lineages that would otherwise require unnecessarily complex descriptions.)
After taking both past and present studies into account, and carefully weighing up the available spectrum of evidence, we conclude that the ITS phylogeny (Fig. 4) is most likely to contain the greatest proportion of accurately reconstructed phylogenetic nodes. The plastid region trnL-F yielded undesirably few informative sites and thus gained poor statistical support across the tree. The morphological matrix incurred exceptionally high homoplasy, apparently encompassing both extensive convergence and occasional character reduction or loss; moreover, the combined total of 13% of cells coded as either polymorphic or unknown further reduced statistical confidence. A topology entirely congruent with our ITS tree between Gymnadenia conopsea s.s., G. odoratissima, G. densiflora and the former genus Nigritella (the latter paired as sisters) was generated by Sun et al. (2015) after adding to ITS sequences data from three low-copy nuclear genes.
Summary: Genus-level circumscription reviewed — Taxonomic literature published during the molecular era has led to the DNA-based classification being followed by some, questioned and selectively rejected by others on the basis of inadequate evidence (a "final solution" was rather inadvisedly demanded by Kropf in Kadereit et al. 2016: 43), or simply ignored without comment by perhaps the majority (reviewed by Bateman 2009, 2012a). In truth, the accumulated evidence supports the molecular re-circumscription of the taper-tubered genera that accommodates 'Nigritella' and 'Pseudorchis' frivaldii within Gymnadenia s.l. and 'Coeloglossum' within Dactylorhiza.
Ongoing opponents of classifications that prioritise DNA data tend to rely on special pleading rather than explicitly proposing their own rule-based logic or directly challenging monophyly as the fundamental classificatory concept (e.g. Devos et al. 2006a; Tyteca et al. 2012; Delforge 2016; Kropf in Kadereit et al. 2016; Perazza 2016). The few attempts to "rescue" Nigritella as a full genus or occasionally to incorporate Pseudorchis into Gymnadenia (e.g. Delforge 2006 — a decision wisely reversed by Delforge 2016) can only be viewed as brave in the light of so much contradictory DNA-based evidence. Even if trees showing the non-nigritellan Gymnadenia species to be monophyletic were eventually to be generated by a future study (as is implied by the unrooted haplotype network recently published by Hedrén et al. 2018), following our explicit rules for deriving classifications from trees (e.g. Bateman & Rudall 2018), the inevitably comparatively short and poorly supported branch separating the two species groups would still require inclusion of 'Nigritella' within Gymnadenia.
The one remaining uncertainty is the phylogenetic position of the monotypic southeast Asian genus Neolindleya, which has been included in at least four molecular studies (Bateman et al. 2003; Inda et al. 2012; Jin et al. 2014; Jin et al. 2017: Fig. 8) and one morphological study (Fig. 5) but has never been placed with >50% bootstrap support; irrespective of the data used, it appears to be intermediate between, and have approximately equal phylogenetic disparities from, Platanthera and Galearis (see also Efimov et al. 2009). On the basis of their multi-gene phylogeny, Jin et al. (2014) argued that Neolindleya should be incorporated into Galearis, but we believe that the genus should be retained despite its monotypic status, at least until multiple sources of evidence are found that together offer much stronger support for one sister-group relationship rather than the other.
Circumscription of, and relationships among, species within genera
Dactylorhiza — Although the most recent taxonomic treatment of the genus Dactylorhiza (Delforge 2016) recognised 60 species as native to Europe plus Asia Minor (though omitted 'Coeloglossum' from the genus), 30 species would be a more scientifically justifiable estimate. Species-level relationships within western European Dactylorhiza s.l. have been explored in several molecular studies (e.g. Hedrén et al. 2001; Bateman et al. 2003; Devos et al. 2006a; Pillon et al. 2007; Hedrén et al. 2008; Hedrén et al. 2011), and those occurring further east in Eurasia have also received informed attention (Hedrén 2001; Hedrén et al. 2007). Given the limited and largely typological sampling of taxa and equally limited sampling of genic regions that characterise representation of Dactylorhiza in the present study, it was unlikely that additional major insights would emerge, and so it proved.
The plastid region trnL-F served only to link two pairs of species already grouped according to classical taxonomy: Dactylorhiza romana with D. sambucina, and D. saccifera with D. fuchsii and the allotetraploid complexes (Fig. 3). However, trnL-F is clearly useful for separating the main phylogenetic groups within the genus and indicates greater disparity among lineages shown as early divergent in the ITS tree. The morphological phylogenetic study similarly offered little resolution, although it did link the Canary Islands endemic D. foliosa with D. fuchsii on the basis of their comparatively thin leaves and trabeculate seed testa. The trabeculate testa is shared with D. majalis — the allotetraploid progeny of members of the D. fuchsii and D. incarnata groups — which is grouped with the latter (its paternal parent) in Fig. 5.
Most of the relationships indicated by the ITS tree receive only modest bootstrap support (Fig. 4). The topology agrees with those of previous studies in showing Dactylorhiza iberica, D. viridis and the D. incarnata group as competing to be the earliest diverging lineage within the genus (reviewed by Bateman & Rudall 2018). Relationships among the remaining species are already well-established, including inheritance of minor modifications of the fuchsii group ribotype by D. majalis and most other allotetraploid taxa.
Gymnadenia — The present study was conceived in the late 1990s primarily to explore species relationships within Gymnadenia s.l. (using Dactylorhiza as a de facto "inner outgroup") and to provide a framework for a parallel population-level study that employed in situ morphometrics in the hope of better circumscribing those species (Bateman & Denholm 2019; R. Bateman, P. Rudall & I. Denholm, unpublished). The study therefore included representatives (typically several) of most species of the genus that have been at least adequately characterised (i.e. excluding several putative species of the former genus Nigritella that are near-identical in both morphology and DNA sequences: reviewed by Teppner & Klein 1985; Teppner 2004; Delforge 2016). Unlike Dactylorhiza, the genus Gymnadenia has not yet been the subject of 21st Century attempts at taxonomic revision, monographers perhaps having been intimidated by accumulating evidence for the presence of several comparatively cryptic species.
The trnL-F phylogeny served largely to distinguish the former genus Nigritella according to its lack of the 9 bp minisatellite, although it also separated the Chinese accession of Gymnadenia orchidis downloaded from the work of Tang et al. (2015) from the remaining species according to several unique SNPs (most of them occurring within the 9 bp minisatellite) plus four indels. Most accessions of all the remaining species shared a single core haplotype, although slightly deviant haplotypes were reported in two accessions of G. conopsea, two of G. densiflora and three of G. odoratissima (Fig. 3); in total, Gymnadenia yielded ten haplotypes (I – X).
As in previous ITS-based studies (Bateman et al. 2003; Gustafsson & Lönn 2003; Bateman et al. 2006; Stark et al. 2011; Efimov 2013; Sun et al. 2015), the resulting trees showed good resolution and taxonomic grouping but only modest bootstrap support in both parsimony and likelihood analyses (Fig. 4). Nonetheless, the improved taxonomic and geographic sampling relative to previous studies allowed some additional, long-standing conundra to be addressed.
In total, 30 ribotypes (I – XXX: Fig. 4) were recovered from the genus Gymnadenia s.l. Stark et al. (2011) reported 35 ribotypes, but unlike us they also included variants that showed ambiguous sites resolved via statistical reconstruction techniques. Our 30 ribotypes boil down to six clades: conopsea plus odoratissima, all available Chinese accessions, borealis, frivaldii, densiflora, and the former genus Nigritella. Gymnadenia encompasses a few widespread low altitude/mid-latitude species (conopsea s.s., densiflora) admixed with montane/high latitude specialists (the remainder). The main taxonomic instability persisting in the genus derives from the southeast European high-altitude specialist G. frivaldii, which has an ITS sequence approximately equally similar to those of G. borealis and G. densiflora plus 'Nigritella'. Setting aside the under-sampled Sino-Himalayan clade, four of the five ITS groups each have a clearly dominant ribotype that is plesiomorphic within the group and hence is presumed to be ancestral (VI, XVII, XIX and XX, respectively, in Fig. 4). The exception is the former genus Nigritella; where by far the more common ribotype (XXX) is derived rather than primitive.
Perhaps the most surprising aspect of both this and previous candidate-gene trees exploring Gymnadenia is their consistent inability to distinguish between the dominantly lowland G. conopsea s.s. and the upland/boreal G. odoratissima. Ribotype VI is most common among individuals of both species, despite the fact that these species are readily distinguished morphologically — they are separated by a disparity of seven characters on our constrained morphology tree (Fig. 7). Most of the remaining ribotypes in the conopsea–odoratissima clade deviate by a single step, although two or three steps separate apparent regional genetic divergences in central and eastern Russia (ribotype I: Efimov 2013) and Germany (ribotypes X and XI: Stark et al. 2011). Three of the samples sequenced for ITS by us that were supposedly members of other species actually yielded ribotypes typical of conopsea and odoratissima and therefore appear to have been mis-identified when collected. One sample originally attributed to "densiflora", collected on our behalf in the Pyrenees, yielded ribotype VIII; we have no knowledge of its morphology, but its flowering time suggests a more likely attribution to conopsea s.s. Two other samples collected on our behalf from the Norwegian coast had a morphology and habitat preference consistent with G. borealis but surprisingly yielded the classic conopsea–odoratissima ribotype VI.
Gustafsson & Sjögren-Gulve (2002) found similarly admixed microsatellites in populations of Gymnadenia conopsea and G. odoratissima in Sweden, although Sun et al. (2015) had more success in distinguishing the two species within 11 mixed-ploidy populations of these species in Switzerland, a result achieved by adding three low-copy nuclear regions to their ITS data. However, Sun et al. (2015) erroneously claimed that their tree showed the two species to be sisters. In fact, their molecular tree shows (admittedly with limited statistical support) G. odoratissima to be monophyletic but originating from within G. conopsea s.s., rendering conopsea paraphyletic. Nonetheless, at least one of their low-copy regions is clearly competent to reliably distinguish (albeit narrowly) between the two species, at least in Switzerland. Huber et al. (2005) reported different spectra of fragrance volatiles, and thus of pollinators, between Swiss populations of the two species.
The next clade of ribotypes to diverge in the ITS tree was confined to southwest China and encompassed all of the Sino-Himalayan taxa analysed by us. Support for this clade also exists in the form of a consistent 1 bp insertion toward the 5' end of the trnS-trnG spacer in the plastid genome (Y. Pillon, pers. comm. 2004). Five putative species of Gymnadenia were recognised in the Flora of China (Chen et al. 2009): long-spurred G. conopsea s.s. and G. orchidis, together with three less well-known short-spurred taxa: G. crassinervis, G. emeiensis and G. bicornis. Traditional taxonomic accounts suggest that G. emeiensis and G. bicornis are typically vegetatively robust, morphologically distinct from the remaining taxa but perhaps not from each other. Unfortunately, we were unable to obtain DNA samples of either putative species, and within this group as a whole, our sampling is insufficient to identify a core ribotype. Our ITS study did, however, show that our two samples of the geographically widespread G. orchidis, which differs from G. conopsea s.s. primarily in its broader leaves, to be paraphyletic relative to three samples of G. crassinervis (Fig. 4). Most perturbing is a sample attributed to G. conopsea by Pillon et al. (2006) but placed here with 97% bootstrap support as the earliest divergent lineage within the Sino-Himalayan clade. It seems likely that plants growing in southeast Asia that closely resemble G. conopsea s.s. in morphology deviate more substantially in ITS sequences, as discussed below for G. densiflora. Viewed together, these results suggest that Sino-Himalayan samples of Gymnadenia have not been consistently identified. More fundamentally, they suggest that circumscription of Gymnadenia species in the region remains far from optimal and is in considerable need of a combined morphometric and population genetic survey.
The most appropriate taxonomic status for Gymnadenia borealis has long been debated (cf. Rose 1991; Lang 2001; Bateman et al. 2003; Bateman et al. 2006), as has its possible confinement to the British Isles (cf. Bateman et al. 2006; Harrap & Harrap 2009; Rankou 2011; Travnicek et al. 2012). Delforge (2006) elected to treat both borealis and densiflora as mere taxonomic varieties in the third edition of his monograph, later illogically elevating borealis but not densiflora to species status in the fourth edition (Delforge 2016). Our five bona fide samples of the species yielded three slightly divergent ribotypes (Fig. 4) and, predictably, a core ribotype that was concentrated in Scotland (XVII). Morphologically the plants resemble miniaturised versions of G. conopsea s.s., although the restriction of G. borealis populations to acid heaths spatially separates them from those of G. conopsea s.s. Thus, the southern English outlier population that we sampled in a Sussex heath yielded a ribotype that was only slightly divergent from the remainder and was clearly attributable to G. borealis. We continue to fail to find bona fide populations of this species in mainland Europe, despite the seemingly similar morphology and habitat preference of some populations in northern Norway (Bjerke & Strann 2009).
Gymnadenia (formerly Pseudorchis) frivaldii, an alpine specialist of wet acid heaths endemic to the Balkans, was the subject of a morphological and molecular study by Bateman et al. (2006) and so need not be discussed in detail here. All four of the northern Greek and Bulgarian samples at our disposal yielded the species-specific ribotype XIX, and this genetic uniformity apparently extends to the morphology of its populations; its distinctive small, pale pink flowers are borne in compact inflorescences on plants that show comparatively little variation in size.
The molecular phylogenetic disparity that separates Gymnadenia densiflora from G. conopsea s.s. (Fig. 4) continues to perplex observers despite their similar morphologies. Substantial differences are evident in both allozyme studies (cf. Scacchi & de Angelis 1989; Soliva & Widmer 1999; Gustafsson 2000) and sequencing data (Gustafsson & Lönn 2003; Bateman et al. 2006; Campbell et al. 2007; Jersáková et al. 2010; Stark et al. 2011; Meekers et al. 2012; Efimov 2013; Saenruen 2014). Gymnadenia densiflora inhabits calcareous to neutral marshes across Europe, and the biochemistry of its scent apparently differs from those of both G. conopsea s.s. and G. odoratissima (Gupta et al. 2014), although the difference in fragrances may nonetheless be insufficient to influence pollinator behaviour (Jersáková et al. 2010). Despite the large number of papers demonstrating a reliable and substantial molecular separation, in the eyes of a few authors, G. densiflora still remains an infraspecific taxon within G. conopsea, either a subspecies (e.g. Lang 2001; Gustafsson & Lönn 2003) or even a mere variety (Kreutz 2004; Bournérias & Prat 2005; Saenruen 2014; Delforge 2006, 2016).
A combination of our own accessions plus GenBank sequences provided us with extensive Europe-wide coverage of this species, yet the ITS clade for densiflora remained stubbornly robust. Most of the deviations from the core ribotype (XX) were slight, although a more interesting and unexpected deviation of four steps distinguished late-flowering plants that were assignable to Gymnadenia densiflora but occurred in both dune-slacks in southern Wales (Kreutz & Lewis 2015) and chalk downland in southern England (cf. Lang 2001; Campbell et al. 2007). Morphologically similar plants occupying dune slacks in the Dutch Frisian Islands were named in rapid succession G. conopsea var. friesica (Kreutz & Lewis 2015) and then G. densiflora var. friesica (Lewis 2015), but have not yet been subjected to either molecular or morphometric study to test whether they represent the same biological entity. However, the flow cytometry profiles of these plants show diploidy and are consistent with G. densiflora (P. Travnicek, pers. comm. 2012).
It is possible that this ITS-delimited taxon — broadly resembling typical marsh-dwelling Gymnadenia densiflora in both morphology (R. Bateman, P. Rudall and I. Denholm, unpublished) and ribotypes (Fig. 4) but tolerant of at least periodically drier soils — could at least partly explain the conundrum presented by a series of reports that G. conopsea s.s. maintains both early-flowering and late-flowering forms. The latter bloom contemporaneously with G. densiflora and reputedly maintain less variation, both morphological and molecular (cf. Gustafsson & Lönn 2003; Marhold et al. 2005; Lönn et al. 2006; Vöth & Sontag 2006; Jersáková et al. 2010; Stark et al. 2011; Saenruen 2014; Gross & Schiestl 2015). Several studies (Gustafsson & Lönn 2003; Stark et al. 2011; Saenruen 2014) have argued that the late-flowering populations of G. conopsea s.s. from Sweden group with G. densiflora in ITS trees, a conclusion that would be in accord with our results from friesica-like populations in England (Fig. 4). However, the converse situation was reported for Czech populations, where the late-flowering tetraploid populations assigned to G. conopsea s.s. have retained the ribotypes of the early-flowering diploid populations (Jersáková et al. 2010). The main weakness of these studies is that only Stark et al. (2011) seriously addressed the morphology of the plants described as late-flowering G. conopsea, and even they scored only nine morphological characters. Early investigations of ploidy suggested that conopsea was predominantly diploid and densiflora tetraploid (e.g. Groll 1965; Jongepierová & Jongepier 1989; Mrkvicka 1993), but more recently this viewpoint has become inverted, densiflora being regarded as predominantly diploid and conopsea as similarly predominantly diploid in western and northern Europe but often tetraploid (with subordinate triploids and pentaploids) further south and east, notably in the Alps and Pyrenees (Marhold et al. 2005; Jersáková et al. 2010; Stark et al. 2011; Trávnicek et al. 2012).
Despite being generally considered to contain approximately 12 species (Hedrén et al. 2000; Kreutz 2004; Teppner 2004; Delforge 2016), the former genus Nigritella yielded only two ribotypes and, in contrast with all other clades within Gymnadenia, by far the most common ribotype (XXX) proved to be derived (Fig. 4). The rarer ribotype (XXIX) appears to be confined to a single species — the diploid, allogamous Pyrenean endemic G. gabasiana. This species is particularly intriguing because its ribotype lacks four of the five synapomorphies possessed by the remaining 'Nigritella' species (only the non-homoplastic C–T transition at position 378 was present in all species analysed). However, when G. gabasiana was assessed for possible inclusion in our morphometric matrix it proved to be identical to the widespread diploid G. rhellicani in all characters that could be scored (sadly, G. gabasiana was omitted from the recent morphometric study by Lorenz & Perazza 2012, which focused on Alpine rather than Pyrenean species). This strong similarity reduces the probability of a recent origin through hybridity analogous to that of the localised apomict Gymnigritella runei in Sweden (Stahlberg 1999; Hedrén et al. 2000) or pentaploid G. buschmanniae in the Italian Alps (Hedrén et al. 2018). It also increases the likelihood that G. gabasiana resembles — and could even be — the ancestor of the remaining 'Nigritella' species.
Interestingly, Gymnadenia gabasiana was the only species that formed a cohesive group in the AFLP analysis of Stahlberg (1999), in which geographic origin influenced inferred relationships among samples more strongly than did taxonomic assignment. Also, Stahlberg placed G. gabasiana as the earliest-diverging species in his UPGMA tree (but see the criticisms of UPGMA clustering outlined below). In contrast, gabasiana failed to form a cohesive group when subjected to allozyme profiling, and it appeared derived rather than primitive in the resulting UPGMA phenograms (Hedrén et al. 2000). Similarly, the haplotype network of Hedrén et al. (2018) nested G. gabasiana (appearing monophyletic in this data-set) within 'Nigritella', their topology suggesting that it diverged later than the autogamous polyploids G. widderi and G. archducis-joannis — taxa that may be conspecific and are highly localised within the Alps and Apennines.
Our failure to detect any differentiation in either ITS or trnL-F among the remaining putative species of 'Nigritella' suggests either over-splitting into species via traditional taxonomic practices, recent diversification, or (as inferred by Hedrén et al.) both. These species maintain a reservoir of morphological character variation (albeit subtle, as evidenced by our morphology trees: Figs 4, 7), often occupy contrasting mountain ranges, present a spectrum of reproductive modes from allogamy to autogamy (including apomixis) that correlate with a range of ploidies and karyotypes (cf. Teppner & Klein 1985; Stahlberg 1999; Hedrén et al. 2000; Lorenz & Perazza 2012; Hedrén et al. 2018). Building upon the excellent taxon molecular foundations recently laid by Hedrén et al. (2018) using haplotypes, ITS sequences and nuclear SSRs for a good spectrum of samples, the former genus Nigritella appears to us to be an ideal subject for a more detailed phylogeographic study of the influence of repeated glaciations on speciation, as discussed in the following section.
Likely causes of speciation within the digitate-tubered clade
The divergence in ITS and plastid regions between Dactylorhiza s.l. and Gymnadenia s.l. is less than that evident between any other genus-level sisters recognised by us within Orchidinae other than the specialised Alpine pairing of Traunsteinera versus Chamorchis. It is therefore unsurprising that in nature intergeneric hybrids have been recorded at F1 level and sometimes beyond between a large percentage of the combinations of species permitted by geographic distributions and ecological preferences (e.g. Oddone et al. 2016; Stace et al. 2015). Artificial breeding experiments (S. Malmgren, pers. comm. 2015; J. Haggar, pers. comm. 2017) have shown both genera to have at most weak post-zygotic isolation mechanisms, as have G. conopsea and G. odoratissima within the genus Gymnadenia (Sun et al. 2015; but see Sletvold et al. 2012b). Hybrids between either Dactylorhiza or Gymnadenia and Pseudorchis are considerably rarer, presumably reflecting the greater molecular divergence of the latter.
Dactylorhiza and Gymnadenia share a chromosomal fusion event that yielded n = 20, a character which is readily coded in a cladistic matrix (Table 3). They also show an unusually strong tendency toward both auto- and especially allopolyploidy, a trend rather than a discrete character and hence less readily scored. It has long been known that the D. majalis complex (whose species richness continues to be much debated) is the allopolyploid product of repeated polyploidisation between D. fuchsii-like and D. incarnata-like parents (Heslop-Harrison 1954; Heslop-Harrison 1968; Hedrén et al. 2008; Paun et al. 2010; Hedrén et al. 2011; Balao et al. 2016). More recently, similar events have been demonstrated in eastern European analogues that include D. cordigera (Hedrén et al. 2007) (Fig. 3). Limited evidence has now accrued for a possible third allopolyploid complex occurring in the Sino-Himalayan region. Autopolyploidy has long been known to characterise D. maculata (Hagerup 1944) and has now been inferred to occur in the earlier-divergent D. viridis (R. Bateman, P. Rudall & I. Denholm, unpublished). Both primary hybrids and allotetraploids reliably place closer to their ovule-parent than their pollen parent in morphological analyses (Fig. 5), suggesting a significant element of epigenetic inheritance in determining the phenotype of the next generation.
Recognition arose only later that similar karyological fluidity pertains within both Gymnadenia s.s. (Marhold et al. 2005; Trávnicek et al. 2012) and the former genus Nigritella (Teppner & Klein 1985; Hedrén et al. 2000; D'Emerico & Grünanger 2001). Extensive surveys across Continental Europe of base genome size in G. conopsea s.s. and G. densiflora by the late Jan Suda and colleagues have revealed a startling complexity of cytotypes that are dominated by diploids, triploids and tetraploids and can extend as far as hexaploids (Trávnicek et al. 2011; Trávnicek et al. 2012). Moreover, Gymnadenia species routinely exhibit the unusual phenomenon of progressively partial endoreplication, duplicating most but not all of the genetic material present in the nucleus in the absence of cell division (Hribova et al. 2016). In contrast, the apparently exclusively British and Irish G. borealis proved to be almost wholly diploid, as did — albeit on the basis of more limited sampling — the Carpathian endemic G. frivaldii and more widespread G. odoratissima (Trávnicek et al. 2012). These observations cast some doubt on recent arguments that polyploidy may confer selective advantage in mixed-ploidy populations of G. conopsea s.s. and G. densiflora (e.g. Jersáková et al. 2010; Gross & Schiestl 2015). Nevertheless, the remarkable lability of ploidy levels within several species of both genera, combined with weak post-zygotic isolation, leaves the clade subject to iterative allopolyploidy of the kind most effectively detailed in the Dactylorhiza majalis complex.
The patterns of morphological diversification contrast between Dactylorhiza s.l. and Gymnadenia s.l. That of Dactylorhiza is more typical of eukaryotic clades in general, being fractal: the early-divergent species D. iberica and D. viridis have accumulated much greater character change than have later-diverging species (Fig. 7), to the extent that D. viridis remains accused by some observers of being better treated as a separate genus (reviewed by Bateman & Rudall 2018). Although the more derived species of Dactylorhiza differ in relatively few characters both morphologically and molecularly, they are readily distinguishable by field botanists with the exception of the allopolyploid complexes. And natural hybridisation occurs readily throughout the genus, wherever biogeography, habitat preference and phenology permit (Eccarius 2016; Stace et al. 2015). Hybrids between allotetraploids and their diploid parents are particularly frequent and are problematic to identify with confidence due to their strong similarity in both phenotype and genotype.
Remarkably, the converse pattern of morphological diversification apparently pertains within Gymnadenia (Fig. 7). Here, it is the earlier divergent species that are more likely to differ only subtly in both morphological and molecular properties, whereas the long-branch clade — the former genus Nigritella — is nested well within Gymnadenia s.l. in our trees. We were surprised that G. odoratissima did not place as sister to 'Nigritella' in our morphological cladistic tree (Fig. 5), as these taxa share the unusual character states of whorled leaves and papillate bract cells (Table 3). Instead, it is G. orchidis that is improbably shown as sister to 'Nigritella', despite the fact that the proximal margins of their present distributions are separated by c. 4,000 km. The ITS-based placement of G. densiflora as sister to 'Nigritella' is more consistent with biogeography and phenology (both taxa flower comparatively late) but is less persuasive in terms of either habitat preference or morphology. Thus, the identity of the sister-species to 'Nigritella' remains in doubt.
The origin of the 'Nigritella' clade has long fascinated one of us as bearing all the likely hallmarks of a saltational morphological speciation event (e.g. Bateman & DiMichele 2002). Specifically, the reduced flower size, simplified labellum and spur, and loss of resupination are features commonly attributed to pseudopeloria (Bateman & Rudall 2006) and would be more easily achieved through genetic or epigenetic suppression of a key developmental gene rather than through gradual acquisition of several smaller changes during the short time-span that has been available to evolution to so radically modify the basic Gymnadenia morphology. The recognition that the diploid G. gabasiana is almost intermediate in ITS sequences between the remainder of 'Nigritella' and G. densiflora (Fig. 4), yet apparently possesses all of the phenotypic features typical of the 'Nigritella' morphology (Figs 5, 7), further reduces the time available for these evolutionary steps to accrue sequentially, and so increases the credibility of the saltational hypothesis.
In addition, the short molecular branch subtending 'Nigritella' raises the possibility that this remarkable phenotype arose in lowland areas during one of the Quaternary glaciations, retreating to montane refugia as the climate ameliorated to temperate conditions. An initially periglacial environment would have largely constrained potential pollinating insects to small-bodied guilds that would have been more likely to be compatible with these small-bodied, small-flowered orchids than with larger-flowered antecedents. We further note that most of the later-derived 'Nigritella' species resulting from allopolyploidy (the majority) indulge in autogamy rather than allogamy (Stahlberg 1999; Hedrén et al. 2000; Claessens & Kleynen 2011; Hedrén et al. 2018).
Setting aside 'Nigritella', the most striking aspect of the relationships inferred within Gymnadenia s.s. is the phylogenetic alternation of species that bear flowers that are (a) medium-sized (G. conopsea, G. orchidis, G. densiflora) versus small and (b) those that bear spurs that are long (the above plus G. borealis) versus short; these phenotypic groups do not form clades (Fig. 7). Reduction in plant and flower size has been sufficiently extensive in G. frivaldii (syn. 'Pseudorchis' frivaldii) to require changes in several characters, resulting in phenotypic convergence with bona fide Pseudorchis (Bateman et al. 2006). We suspect that the larger flowers were ancestral to the genus, which subsequently experienced iterative bouts of paedomorphic heterochrony to produce the smaller-flowered taxa, culminating in the more profound phenotypic shifts evident in G. frivaldii and the former genus Nigritella (Box et al. 2008; Bateman 2012b). If so, morphological convergence would be confined to the smaller-flowered species of Gymnadenia, the larger-flowered species more closely representing the common ancestor of the genus (Fig. 7). In order for this hypothesis of iterative paedomorphosis to be explored more effectively, and to tease out the significance of the contrasting habitat preferences of the controversial western European species G. conopsea s.s., G. densiflora and G. borealis, continuous morphological characters need to be added to the more discrete characters already scored. This approach would transfer the analytical emphasis to the realm of morphometric ordination rather than phylogeny reconstruction. Such a study is currently in preparation, building on the morphometric comparison of G. conopsea and G. densiflora presented by Stark et al. (2011) but based on a much wider range of characters and comparing all of the European species of Gymnadenia s.s. (Bateman and Denholm 2019; R. Bateman, P. Rudall & I. Denholm, unpublished).
Gymnadenia conopsea s.s. was determined to be a mycorrhizal generalist by Stark et al. (2009), whereas Tesitelová et al. (2013) detected divergence between the mycorrhizal associates of both seeds and adult plants of diploid versus tetraploid individuals collectively assigned by them to G. conopsea s.s. Numerous authors have stated that G. conopsea s.s., like most orchids that offer substantial nectar rewards, is a generalist with regard to pollination (e.g. Vöth 2000; Meyer et al. 2007; Meekers et al. 2012; Sletvold et al. 2012a). Claessens & Kleynen (2011) listed 40 species of Coleoptera, Diptera, Hymenoptera and especially Lepidoptera that are implicated as its pollinators (although it is unlikely that all of these observations actually pertain only to G. conopsea s.s.). However, despite this broad spectrum of pollinators, Sun et al. (2015) provided evidence that there is little if any gene flow affecting G. conopsea where it co-occurs with G. odoratissima, at least within their Swiss study populations. Interest will no doubt be maintained in the pronounced fragrances of these plants, not least because recent research showed that diurnal and nocturnal emissions differ in quantitative composition and that nocturnal divergence between populations is greater than diurnal divergence (Chapurlat et al. 2018).
As with Dactylorhiza, most species of Gymnadenia that come into regular contact have proven capable of producing occasional natural hybrids to at least F1 generation (although these are difficult to identify morphologically when the parents are phenotypically similar, as in the case of G. conopsea s.s. × G. densiflora: reviewed by Bateman et al. 2006); moreover, Gymnadenia species cross readily in cultivation. For example, G. conopsea s.s. and G. odoratissima can produce natural hybrid swarms even in undisturbed habitats (R. Bateman, unpublished) and are easily artificially crossed as far as an F3 generation (although the F3 plants appear surprisingly unappealing to potential pollinating insects: S. Malmgren, pers. comm. 2015). Indeed, Sun et al. (2015) showed that the two species owe their restricted gene flow largely to pre-zygotic pollinator choice, which is assumed to be influenced primarily by demonstrated differences in the composition of their respective scent cocktails. On the other hand, frequent hybridisation between species of Gymnadenia and those of Dactylorhiza indicates that at least some pollinators remain blissfully ignorant of the behaviour that they are required by theory to exhibit (reviewed by Bateman et al. 2017).
Overall, a striking diversity of evolutionary mechanisms are implicated as having contributed appreciably to the diversity of both characters and species evident within the digitate-tubered clade (Bateman 2009; Bateman 2012b). They include allopolyploidy, autopolyploidy, mutationally driven lineage divergence ('dichotomous saltation' sensu Bateman & DiMichele 2002), geographical isolation, diversification of ecotypes into contrasting habitats, autogamy, and perhaps even pollinator switching.
Conclusions
-
1)
Several molecular phylogenetic studies based on Sanger sequencing have provided an optimal circumscription of genera within the taper-tubered clade of subtribe Orchidinae (although we remain unable to confidently incorporate the monotypic genus Neolindleya into either Galearis s.l. or Platanthera s.l.). However, the approach has been less successful at determining relationships among the genera. The best-supported phylogenetic nodes are the focus of this paper — those subtending the digitate-tubered clade (Dactylorhiza s.l. plus Gymnadenia s.l.) and the taper-tubered clade (the two digitate-tubered genera plus Platanthera, Galearis and Neolindleya). Ironically, it is the genus-level circumscriptions that continue to attract greatest criticism (reviewed by Bateman 2012a), despite the fact that they are the most conclusive result of two decades of molecular phylogenetic study pursued in a rigorous conceptual framework by multiple research groups.
-
2)
Intriguingly, the application of parsimony to our morphological cladistic matrix reproduced a topology (albeit highly unstable) that closely resembled the most common genus-level classification of Orchidinae achieved through traditional authoritarian taxonomy, prior to the advent of molecular phylogenetics. The pre-molecular classification would, from first principles, have been predicted to have been more closely mirrored by phenetic analyses of our matrix, yet the equivalent NJ-based and especially UPGMA-based morphological topologies diverge more strongly from molecular topologies.
-
3)
The contrasting phylogenetic topologies obtained here between nuclear ribosomal, plastid and morphological cladistic matrices challenge the wisdom of the now near-ubiquitous practice of routinely combining such data sets, given that they evolve within highly contrasting milieux. Similar conclusions have been reached following studies of other groups of orchids (e.g. van de Niet & Linder 2008; Sramkó et al. 2014; Tang et al. 2015; Pérez et al. 2016). Our results also emphasise the crucial importance of intensive sampling of species across the groups of interest. It remains to be seen whether topologies based on matrices obtained through next-generation sequencing technologies (reviewed by Olson et al. 2016) will offer the substantially greater stability that some commentators anticipate (e.g. Kropf in Kadereit et al. 2016; Pellegrino & Cozzolino 2016); our initial results suggest that only modest improvements are likely (Bateman et al. 2017; G. Sramkó, R. Bateman & O. Paun, unpublished; R. Bateman, P. Rudall & O. Pérez, unpublished). Unfortunately, morphological cladistic matrices — the foundation of the phylogenetics revolution and still a valuable component of any genuinely integrated comparative study — are rarely produced today, having become seriously under-valued by the research community.
-
4)
We offer further support for earlier pre-molecular (e.g. Vermeulen 1947; Heslop-Harrison 1954, Vermeulen 1977) and molecular phylogenetic (e.g. Bateman et al. 1997; Pridgeon et al. 1997; Bateman et al. 2006) recognition that tapered tubers (those expanded into one or more distal roots) delimit the broader clade, and that apically divided digitate tubers plus a chromosomal fusion event (n = 21 > 20) plus a predisposition to polyploidy together delimit the digitate-tubered clade.
-
5)
Mapping of 51 morphological cladistic characters across (i.e. constrained to) an ITS-based molecular topology increased levels of homoplasy in an already highly homoplastic morphological matrix by a further 20%, indicating an exceptionally high degree of evolutionary lability within the digitate-tubered clade that reflects convergence and so-called 'losses' of features, although in truth, paedomorphic reduction is a more frequent phenomenon than complete loss.
-
6)
NrITS remains the best single region of choice for classic phylogenetic purposes approximating the genus level, due to its high mutation rate and coalescence properties (e.g. Hein et al. 2004; Bateman 2018). Ironically, these properties are often misrepresented as a negative feature, the term 'concerted evolution' often being applied in a pejorative context. Nonetheless, the failure of ITS to distinguish between the morphologically distinct species Gymnadenia conopsea s.s. and G. odoratissima demonstrates that at least a small minority of bona fide orchid species exist within the "genetic divergence lag phase" of Bateman (2016), thereby showing that even ITS is not a panacea for species delimitation. The present study will be used as a framework for a future paper comparing in detail the morphometric properties of all species of Gymnadenia other than those from southeast Asia (R. Bateman, P. Rudall & I. Denholm, unpublished), with the aim of better characterising these problematic species in a way more useful to typical field botanists.
-
7)
One clear message to emerge from this study (yet again) is the great desirability, when circumscribing species and/or attempting to understand the underlying speciation mechanisms, of combining genetic studies with morphometric surveys of the same study populations. Most of the molecular studies cited in this paper (including many of those published by the present authors) have relied entirely on molecular data for their conclusions, yet it is phenotypes rather than genotypes that dictate any form of interaction between organisms. For example, the failure to collect morphometric data in most studies (the notable exception being Stark et al. 2011) when studying the relationship between Gymnadenia conopsea (diploid), G. conopsea (tetraploid) and G. densiflora (reputedly mostly diploid) has left the significance of studies of these three (or more?) biological entities virtually indecipherable — it is not clear exactly which phenotypes have been studied (and would not be even if herbarium vouchers were available for each plant studied — they are inevitably subject to post-mortem changes in morphometric parameters).
-
8)
Dactylorhiza s.l. has become a model system for studying the adaptive and/or epigenetic consequences of ploidy change (Paun et al. 2010; Paun et al. 2011; Balao et al. 2016; Balao et al. 2017), while Gymnadenia s.s. has provided valuable information about the phylogeography of ploidy change (Trávnicek et al. 2011; Trávnicek et al. 2012) and the attraction of pollinators to orchid species that offer substantial nectar rewards (Huber et al. 2005; Lönn et al. 2006; Jersaková et al. 2010; Sletvold & Agren 2011; Sletvold et al. 2012a; Gupta et al. 2014; Gijbels et al. 2015; Gross & Schiestl 2015; Sun et al. 2015). Within Gymnadenia s.l., the radically morphologically divergent former genus Nigritella remains a strong candidate for having evolved from within Gymnadenia s.s. by saltational rather than gradual evolutionary mechanisms (Bateman & DiMichele 2002) and should be a prime target for future evolutionary-developmental genetic study. The ability of the genus to diversify into highly contrasting ecotypes (cf. G. conopsea s.s., G. densiflora and G. borealis in the British Isles) also merits investigation.
References
Balao, F., Tannhäuser, M., Lorenzo, M. T., Hedrén, M. & Paun, O. (2016). Genetic differentiation and admixture between sibling allopolyploids in the Dactylorhiza majalis complex. Heredity 116: 351 – 361.
Balao, F., Trucchi, E., Wolfe, T. M., Hao, B.-H., Lorenzo, M. T., Baar, J., Sedman, L., Kosiol, C., Amman, F., Chase, M. W., Hedrén, M. W. & Paun, O. (2017). Adaptive sequence evolution is driven by biotic stress in a pair of orchid species (Dactylorhiza) with distinct ecological optima. Molec. Ecol. 26: 3649 – 3662.
Barone Lumaga, M. R., Cozzolino, S. & Kocyan, A. (2006). Exine micromorphology of Orchidinae (Orchidoideae, Orchidaceae): phylogenetic constraints or ecological influences? Ann. Bot. 98: 237 – 244.
Bateman, R. M. (2009). Evolutionary classification of European orchids: the crucial importance of maximising explicit evidence and minimising authoritarian speculation. J. Eur. Orch. 41: 243 – 318.
Bateman, R. M. (2012a). Circumscribing genera in the European orchid flora: a subjective critique of recent contributions. Ber. Arbeitskr. Heim. Orch. 29: 92 – 124.
Bateman, R. M. (2012b). Circumscribing species in the European orchid flora: multiple datasets interpreted in the context of speciation mechanisms. Ber. Arbeitskr. Heim. Orch. 29: 160 – 212.
Bateman, R. M. (2016). Après le déluge: ubiquitous field barcoding should drive 21st century taxonomy (Ch. 6). In: P. D. Olson, J. Hughes & J. A. Cotton (eds), Next generation systematics, pp. 123 – 153. Cambridge University Press, Cambridge.
Bateman, R. M. (2018). Two bees or not two bees? An overview of Ophrys systematics. Ber. Arbeitskr. Heim. Orch. 35: 5 – 46.
& Denholm, I. (2019). Can the third atlas more effectively map the near-cryptic Fragrant Orchids of Britain and Ireland? BSBI News 140 (in press: c. 4 pp.)
Bateman, R. M. & DiMichele, W. A. (2002). Generating and filtering major phenotypic novelties: neoGoldschmidtian saltation revisited (Ch. 7). In: Q. C. B. Cronk, R. M. Bateman & J. A. Hawkins (eds), Developmental genetics and plant evolution, pp. 109 – 159. Taylor & Francis, London.
Bateman, R. M., Hollingsworth, P. M., Preston, J., Luo, Y.-B., Pridgeon, A. M. & Chase, M. W. (2003). Molecular phylogenetics and evolution of Orchidinae and selected Habenariinae (Orchidaceae). Bot. J. Linn. Soc. 142: 1 – 40.
Bateman, R. M., James, K. E., Luo, Y.-B., Lauri, R. K., Fulcher, T., Cribb, P. L. & Chase, M. W. (2009). Molecular phylogenetics of the Platanthera clade (Orchidaceae: Orchidinae) prompts expansion of the generic limits of Galearis and Platanthera. Ann. Bot. 104: 431 – 445.
Bateman, R. M., Murphy, A. R. M. & Tattersall, B. G. (2017). × Dactylodenia lacerta R. M. Bateman & Tattersall: a morphologically cryptic hybrid orchid new to science from the Lizard Peninsula, Cornwall. New J. Bot. 7: 64 – 77.
Bateman, R. M., Pridgeon, A. M. & Chase, M. W. (1997). Phylogenetics of subtribe Orchidinae (Orchidoideae, Orchidaceae) based on nuclear ITS sequences. 2. Infrageneric relationships and taxonomic revision to achieve monophyly of Orchis sensu stricto. Lindleyana 12: 113 – 141.
Bateman, R. M. & Rudall, P. J. (2006). The Good, the Bad and the Ugly: using naturally occurring terata to distinguish the possible from the impossible in orchid floral evolution. Aliso (Monocot Spec) 22: 481 – 496.
Bateman, R. M. & Rudall, P. J. (2018). Clarified relationship between Dactylorhiza viridis and Dactylorhiza iberica renders obsolete the former genus Coeloglossum. Kew Bull. 73(21): 4. https://doi.org/10.1007/S12225-017-9728-Z.
Bateman, R. M., Rudall, P. J. & James, K. E. (2006). Phylogenetic context, generic affinities and evolutionary origin of the enigmatic Balkan orchid Gymnadenia frivaldii Hampe ex Griseb. Taxon 55: 107 – 118.
Bateman, R. M., Sramkó, G. & Paun, O. (2018). Integrating restriction site-associated DNA sequencing (RAD-seq) with morphological cladistic analysis clarifies evolutionary relationships among major species groups of bee orchids. Ann. Bot. 121: 85 – 105.
Bateman, R. M., Sramkó, G. & Rudall, P. J. (2015). Floral miniaturisation and autogamy in boreal-arctic plants are epitomised by Iceland’s most frequent orchid, Platanthera hyperborea. PeerJ 3([45 pp.]): e894.
Bell, A. K., Roberts, D. L., Hawkins, J. A., Rudall, P. J., Box, M. S. & Bateman, R. M. (2009). Comparative morphology of nectariferous and nectarless labellar spurs in selected clades of subtribe Orchidinae (Orchidaceae). Bot. J. Linn. Soc. 160: 369 – 387.
Bellusci, F., Pellegrino, G., Palermo, A. M. & Musacchio, A. (2008). Phylogenetic relationships in the orchid genus Serapias L. based on noncoding regions of the chloroplast genome. Molec. Phylogen. Evol. 47: 986 – 991.
Bournérias, M. & Prat, D. (eds) (2005). Les orchidées de France, Belgique et Luxembourg, 2nd ed Biotope, Mezé.
Box, M. S., Bateman, R. M., Glover, B. J. & Rudall, P. J. (2008). Floral ontogenetic evidence of repeated speciation via paedomorphosis in subtribe Orchidinae (Orchidaceae). Bot. J. Linn. Soc. 127: 429 – 454.
Bjerke, J. W. & Strann, K.-B. (2009). Orkideen tettbrudespore Gymnadenia densiflora i Nord-Norge. Blyttia 67: 126 – 133.
Breitkopf, H., Onstein, R. E., Cafasso, D., Schlüter, P. M. & Cozzolino, S. (2015). Multiple shifts to different pollinators fuelled rapid diversification in sexually deceptive Ophrys orchids. New Phytol. 207: 377 – 386.
Campbell, V. V., Rowe, G., Beebee, T. J. C. & Hutchings, M. J. (2007). Genetic differentiation amongst fragrant orchids (Gymnadenia conopsea s.l.) in the British Isles. Bot. J. Linn. Soc. 155: 349 – 360.
Chapurlat, E., Anderson, J., Agren, J., Friberg, M. & Sletvold, N. (2018). Diel pattern of floral scent emission matches the relative importance of diurnal and nocturnal pollinators in populations of Gymnadenia conopsea. Ann. Bot. 121: 711 – 721.
Chen, X., Gale, S. W. & Cribb, P. J. (2009). 38. Gymnadenia. In: Z. Wu, P. H. Raven & D. Hond (eds), Flora of China Vol. 25. Missouri Botanical Garden, St Louis, Missouri.
Claessens, J. & Kleynen, J. (2011). The flower of the European orchid: form and function. Published by the authors, Voerendaal, Netherlands.
Delforge, P. (2006). Orchids of Europe, North Africa and the Middle East. A. & C. Black, London.
Delforge, P. (2016). Orchidées d'Europe d'Afrique du Nord et du Proche-Orient. Delachaux & Niestlé, Paris.
D'Emerico, S. & Grünanger, P. (2001). Giemsa C-banding in some Gymnadenia species and in Chamorchis alpina from Dolomites (Italy). J. Eur. Orch. 33: 405 – 414.
Devey, D. S., Bateman, R. M., Fay, M. F. & Hawkins, J. A. (2008). Friends or relatives? Phylogenetics and species delimitation in the controversial European orchid genus Ophrys. Ann. Bot. 101: 385 – 402.
Devos, N., Raspé, O., Jacquemart, A.-L. & Tyteca, D. (2006b). On the monophyly of Dactylorhiza Necker ex Nevski (Orchidaceae): is Coeloglossum (L.) Hartman a Dactylorhiza? Bot. J. Linn. Soc. 162: 261 – 269.
Devos, N., Raspé, O., Oh, S.-H., Tyteca, D. & Jacquemart, A.-L. (2006a). The evolution of Dactylorhiza (Orchidaceae) allotetraploid complex: insights from nrDNA sequences and cpDNA PCR-RFLP data. Molec. Phylogen. Evol. 38: 767 – 778.
Doyle, J. & Doyle, J. (1990). A rapid total DNA preparation procedure for fresh plant tissue. Focus 12: 13 – 15.
Eccarius, W. (2016). Die Orchideengattung Dactylorhiza. Published by the author, Bürgel, Germany.
Efimov, P. G. (2013). Sibling species of fragrant orchids (Gymnadenia: Orchidaceae: Magnoliophyta) in Russia. Russian J. Genet. 49: 299 – 309.
Efimov, P. G., Lauri, R. K. & Bateman, R. M. (2009). Neolindleya Kraenzl. (Orchidaceae), an enigmatic and largely overlooked autogamous genus from temperate East Asia. Kew Bull. 64: 661 – 671.
Gamarra, R., Galán, P., Herrera, I. & Ortúñez, E. (2008). Seed micromorphology supports the splitting of Limnorchis from Platanthera (Orchidaceae). Nordic J. Bot. 26: 61 – 65.
Gamarra, R., Galán, P., Pedersen, H. A., Ortúnez, E. & Sanz, E. (2015). Seed micromorphology in Dactylorhiza Necker ex Nevski (Orchidaceae) and allied genera. Turkish J. Bot. 39: 298 – 309.
Gijbels, P., Ceulemans, T., Van den Ende, W. & Honnay, O. (2015). Experimental fertilisation increases amino acid content in floral nectar, fruit set and degree of selfing in the orchid Gymnadenia conopsea. Oecologia 179: 785 – 795.
Groll, M. (1965). Fruchtansatz, Bestäubung und Merkmalsanalyse bei diploiden und polyploiden Sippen von Dactylorchis (Orchis) maculata und Gymnadenia conopsea. Österr. Bot. Zeit. 112: 657 – 700.
Gross, K. & Schiestl, F. P. (2015). Are tetraploids more successful? Floral signals, reproductive success and floral isolation in mixed-ploidy populations of a terrestrial orchid. Ann. Bot. 115: 263 – 273.
Gupta, A. K., Schauvinhold, I., Pickersky, E. & Schiestl, F. P. (2014). Eugenol synthase genes in floral scent variation in Gymnadenia species. Funct. & Integr. Genom. 14: 779 – 788.
Gustafsson, S. (2000). Patterns of genetic variation in the fragrant orchid (Gymnadenia conopsea). Molec. Ecol. 9: 1863 – 1872.
Gustafsson, S. & Lönn, M. (2003). Genetic differentiation and habitat preference of flowering-time variants within Gymnadenia conopsea. Heredity 91: 284 – 292.
Gustafsson, S. & Sjögren-Gulve, P. (2002). Genetic diversity in the rare orchid, Gymnadenia odoratissima and a comparison with the more common congener, G. conopsea. Conserv. Genet. 3: 225 – 234.
Hagerup, O. (1944). On fertilisation, polyploidy and haploidy in Orchis maculata L. sens. lat. Dansk Bot Arch. 11: 1 – 25.
Hapeman, J. R. & Inoue, K. (1997). Plant–pollinator interactions and floral radiation in Platanthera (Orchidaceae). In: T. J. Givnish & K. J. Sytsma (eds), Molecular evolution and adaptive radiation, pp. 433 – 454. Cambridge University Press, Cambridge.
Harrap, A. & Harrap, S. (2009). Orchids of Britain and Ireland. A. & C. Black, London.
Hedrén, M. (2001). Systematics of the Dactylorhiza euxina/incarnata/maculata polyploid complex (Orchidaceae) in Turkey: evidence from allozyme data. Plant Syst. Evol. 229: 23 – 44.
Hedrén, M., Fay, M. & Chase, M. W. (2001). Amplified fragment length polymorphisms (AFLP) reveal details of polyploid evolution in Dactylorhiza (Orchidaceae). Amer. J. Bot. 88: 1868 – 1880.
Hedrén, M., Klein, E. & Teppner, H. (2000). Evolution of polyploids in the European orchid genus Nigritella: evidence from allozyme data. Phyton A 40: 239 – 275.
Hedrén, M., Lorenz, R., Teppner H., Dolinar, B., Giotta, C., Griebl, N., Hansson, S., Heidtke, U., Klein, E., Perazza, G., Stahlberg, D. & Surina, B. (2018). Evolution and systematics of polyploid Nigritella (Orchidaceae). Nordic J. Bot. 36: 1 – 32.
Hedrén, M., Nordström, S. & Bateman, R. M. (2011). Plastid and nuclear DNA marker data support the recognition of four tetraploid marsh orchids (Dactylorhiza majalis s.l., Orchidaceae) in Britain and Ireland. Biol. J. Linn. Soc. 104: 107 – 128 (+ 4 electronic appendices).
Hedrén, M., Nordström, S., Hovmalm, H. A. P., Pedersen, H. A. & Hansson, S. (2007). Patterns of polyploidy evolution in Greek marsh orchids (Dactylorhiza; Orchidaceae) as revealed by allozymes, AFLPs, and plastid DNA data. Amer. J. Bot. 94: 1205 – 1218.
Hedrén, M., Nordström, S. & Ståhlberg, D. (2008). Polyploid evolution and plastid DNA variation in the Dactylorhiza incarnata/maculata complex (Orchidaceae) in Scandinavia. Molec. Ecol. 17: 5075 – 5091.
Hein, J., Schierup, M. & Wiuf, C. (2004). Gene genealogies, variation and evolution: a primer in coalescent theory. Oxford University Press, Oxford.
Heslop-Harrison, J. (1954). A synopsis of the dactylorchids of the British Isles. Ber. Geobot. Forsch. Rübel 1953: 53 – 82.
____ (1968). Genetic system and ecological habit as factors in dactylorchid variation. Jber. Naturwiss. Ver. Wuppertal 21+22: 20 – 27
Hribova, E., Holusova, K., Travnicek, P., Petrovska, B., Ponert, J., Simkova, H., Kubatova, B., Jersakova, J., Curn, V., Suda, J., Dolezel, J. & Vrana, J. (2016). The enigma of progressively partial endoreplication: new insights provided by flow cytometry and next-generation sequencing. Genome Biol. Evol. 8: 1996 – 2005.
Huber, F. K., Kaiser, R., Sauter, W. & Schiestl, F. P. (2005). Floral scent emission and pollinator attraction in two species of Gymnadenia (Orchidaceae). Oecologia 142: 564 – 575.
Inda, L. A., Pimental, M. & Chase, M. W. (2012). Phylogenetics of tribe Orchideae (Orchidaceae: Orchidoideae): based on combined DNA matrices: inferences regarding timing of diversification and evolution of pollination syndromes. Ann. Bot. 110: 71 – 90.
Jersáková, J., Castro, S., Sonk, N., Milchreit, K., Schödelbauerová, I., Tolasch, T. & Dötterl, S. (2010). Absence of pollinator-mediated premating barriers in mixed-ploidy populations of Gymnadenia conopsea s.l. (Orchidaceae). Evol. Ecol. 24: 1199 – 1218.
Jin, X. H., Li, D. Z., Xiang, X. G., Lai, Y. J. & Shi, X. C. (2012). Nujiangia (Orchidaceae: Orchideae): a new genus from the Himalayas. J. Syst. Evol. 50: 64 – 71.
Jin, X. H., Schuitemann, A., Chase, M. W., Li, J.-W., Chung, S.-W., Hsu, T.-C. & Jin, X.-H. (2017). Phylogenetics of subtribe Orchidinae s.l. (Orchidaceae; Orchidoideae) based on seven markers (plastid matK, psaB, rbcL, trnL-F, trnH-psba, and nuclear nrITS, Xdh): implications for generic delimitation. BMC Plant Biol. 17: 222 [14 pp.].
Jin, X. H., Schuiteman, A., Li D.-Z., Xiang, X.-G., Huang, W.-C., Li, J.-W. & Huang, L.-Q. (2014). Molecular systematics of subtribe Orchidinae and Asian taxa of Habenariinae (Orchideae, Orchidaceae) based on plastid matK, rbcL and nuclear ITS. Molec. Phylogen. Evol. 77: 41 – 53.
Jongepierova, I. & Jongepier, J. W. (1989). Study on the taxonomy of Gymnadenia conopsea in Czechoslovakia: a preliminary report. Mém. Soc. Roy. Bot. Belges 11: 123 – 132.
Kreutz, C. A. J. (2004). Kompendium der Europäischen Orchideen. Published by the author, Landgraaf.
Kreutz, C. A. J. & Lewis, L. (2015). Neotypification and distribution of Gymnadenia conopsea (L.) R. Brown var. friesica Schechter. J. Eur. Orch. 47: 239 – 249.
Kropf, M. (2016). Orchidaceae. Pp. 43 – 45. In: J. Kadereit and 21 co-authors, What changes are needed to render all genera of the German flora monophyletic? Willdenowia 46: 39 – 91.
Landwehr, J. (1977). Wilde Orchideeën van Europa. Published by the author, Amsterdam.
Lang, D. (2001). Wild orchids of Sussex. Pomegranate Press, Lewes.
Lewis, L. (2015). Reclassification of Gymnadenia conopsea var. friesica as a variety of Gymnadenia densiflora. J. Eur. Orch. 46: 293 – 302.
Lönn, M., Alexandersson, R. & Gustafsson, S. (2006). Hybrids and fruit set in a mixed flowering-time population of Gymnadenia conopsea (Orchidaceae). Hereditas 143: 222 – 228.
Lorenz, R. & Perazza, G. (2012). Beiträge zur Kenntnis der Blütenmorphologie der Artengruppe Nigritella miniata s.l. (Orchidaceae) in der Ostalpen. Gredleriana 12: 67 – 146.
Luer, C. (1975). The native orchids of the United States and Canada, excluding Florida. New York Botanical Garden, New York.
Luo, Y.-B. & Chen, S.-C. (2000). The floral morphology and ontogeny of some Chinese representatives of orchid subtribe Orchidinae. Bot. J. Linn. Soc. 134: 529 – 548.
Maddison, D. R. & Maddison, W. P. (2002). MacClade 4.05. Sinauer, Sunderland, Massachusetts.
Marhold, K., Jongepierová, I., Krahulcová, A. & Kučera, J. (2005). Morphological and karyological differentiation of Gymnadenia densiflora and G. conopsea in the Czech Republic and Slovakia. Preslia 77: 159 – 176.
Meekers, T., Hutchings, M. J., Honnay, O. & Jacquemyn, H. (2012). Biological flora of the British Isles: Gymnadenia conopsea s.l. J. Ecol. 100: 1269 – 1288.
Meyer, B., Kröger, J. & Steffan-Dewenter, I. (2007). Contribution of diurnal and nocturnal pollinators to the reproductive success of the orchid species Gymnadenia conopsea. Entomol. Genet. 30: 299 – 300.
Mrkvička, A. C. (1993). Statistische Untersuchungen an Gymnadenia conopsea (L.) R. Br. s.l. Mitt. Arb. Heim. Orch. Baden-Württem. 25: 361 – 367.
Oddone, N., Andreoli, M. V. & Casabianca, A. (2016). New genus subdivisions and taxonomic revision of intergeneric hybrids in genera Dactylorhiza, Gymnadenia, Pseudorchis and in nothogenera ×Pseudorhiza, ×Pseudadenia and ×Dactylodenia (Orchidaceae). GIROS Orch. Spont. d'Eur. 59: 347 – 377.
Olson, P. D., Hughes, J. & Cotton, J. A., eds (2016). Next generation systematics. Systematics Association Special Volume 85. Cambridge University Press, Cambridge.
Paun, O., Bateman, R. M., Fay, M. F., Hedrén, M., Civeyrel, L. & Chase, M. W. (2010). Stable epigenetic effects impact evolution and adaptation in allopolyploid orchids. Molec. Biol. Evol. 27: 2465 – 2473.
Paun, O., Bateman, R. M., Luna, J. A., Moat, J., Fay, M. F. & Chase, M. W. (2011). Gene expression alteration and ecological divergence in sibling allopolyploids of Dactylorhiza (Orchidaceae). BMC Evol Biol. 11: 113 [14 pp.].
Pellegrino, G. & Cozzolino, S. (2016). Sistemaica e tassonomia. In: G. I. R. O. S. Committee (ed.), Orchidee d'Italia (edn 2), pp. 47 – 54. Il Castello, Milan.
Perazza, G. (2016). Gymnadenia, Nigritella. In: GIROS Committee (ed.), Orchidee d'Italia (edn 2), pp. 90 – 102. Il Castello, Milan.
Pérez, O., Balbuena, J. & Gottschling, M. (2016). Rumbling orchids: how to assess divergent evolution between chloroplast endosymbionts and their nuclear host. Syst. Biol. 65: 51 – 65.
Pillon, Y., Fay, M. F., Hedrén, M., Bateman, R. M., Devey, D., Shipunov, A., van der Bank, M. & Chase, M. W. (2007). Insights into the evolution and biogeography of Western European species complexes in Dactylorhiza (Orchidaceae). Taxon 56: 1185 – 1208.
Pillon, Y., Fay, M. F., Shipunov, A. B. & Chase, M. W. (2006). Species diversity versus phylogenetic diversity: a practical study in the taxonomically difficult genus Dactylorhiza (Orchidaceae). Biol. Conserv. 129: 4 – 13.
Pornon, A., Escaravage, N., Burrus, M., Holota, H., Khimoun, A., Mariette, J., Pellizzari, C., Iribar, A., Etienne, R., Taberlet, P., Vidal, M., Winterton, P., Zinger, L. & Andalo, C. (2016). Using metabarcoding to reveal and quantify plant–pollinator interactions. Nature Sci. Rep. 6: e27282.
Pridgeon, A. M., Bateman, R. M., Cox, A. V., Hapeman, J. R. & Chase, M. W. (1997). Phylogenetics of subtribe Orchidinae (Orchidoideae, Orchidaceae) based on nuclear ITS sequences. 1. Intergeneric relationships and polyphyly of Orchis sensu lato. Lindleyana 12: 89 – 109.
Rankou, H. (2011). Gymnadenia borealis. The IUCN Red List of threatened species 2011: e.T175941A7149754
Rose, F. (1991). A new subspecies of Gymnadenia conopsea (L.) R. Br. Watsonia 18: 319 – 320.
Saenruen, A. (2014). Molecular systematics of the Gymnadenia conopsea complex. In: Masters thesis. Lund University, Sweden.
Scacchi, R. & de Angelis, G. (1989). Isoenzyme polymorphism in Gymnadenia conopsea and its inferences for systematics within this species. Biochem. Syst. Ecol. 17: 25 – 33.
Simmons, M. P. & Ochoterena, H. (2000). Gaps as characters in sequence-based phylogenetic analysis. Syst. Biol. 49: 369 – 381.
Sletvold, N. & Agren, J. (2011). Among-population variation in costs of reproduction in the long-lived orchid Gymnadenia conopsea: an experimental study. Oecologia 167: 461 – 468.
Sletvold, N., Grindeland, J. M., Zu, P. & Agren, J. (2012b). Strong inbreeding depression and local outbreeding depression in the rewarding orchid Gymnadenia conopsea. Conserv. Genet. 13: 1305 – 1315.
Sletvold, N., Trunschke, J., Wimmergren, C. & Agren, J. (2012a). Separating selection by diurnal and nocturnal pollinators on floral display and spur length in Gymnadenia conopsea. Ecology 93: 1880 – 1891.
Soininen, E. M., Valentoni, A., Coissac, E., Miquel, C., Gielly, L., Brochmann, C., Brysting, A. K., Sonstebo, J. H., Ims, R. A., Yoccoz, M. G. & Taberlet, P. (2009). Analysing diet of small herbivores: the efficiency of DNA barcoding coupled with high-throughput pyrosequencing for deciphering the composition of complex plant mixtures. Front. Zool. 6: e16.
Soliva, M. & Widmer, A. (1999). Genetic and floral divergence among sympatric populations of Gymnadenia conopsea s.l. (Orchidaceae) with different flowering phenology. Int. J. Pl. Sci. 160: 897 – 905.
Soliva, M. & Widmer, A. (2003). Gene flow across species boundaries in sympatric, sexually deceptive Ophrys (Orchidaceae) species. Evolution 57: 2252 – 2261.
Sonkoly, J., Votkó, A. E., Tökölyi, J., Török, P., Srankó, G., Illyés, Z. & Molnár, A. V. (2016). Higher seed number compensates for lower fruit set in deceptive orchids. J. Ecol. 104: 343 – 351.
Sramkó, G., Molnár, A. V., Hawkins, J. A. & Bateman, R. M. (2014). Molecular phylogenetics and evolution of the Eurasiatic orchid genus Himantoglossum s.l. Ann. Bot. 114: 1609 – 1626.
Stace, C. A., Preston, C. D. & Pearman, D. A. (2015). Hybrid flora of the British Isles. BSBI, Bristol.
Stahlberg, D. (1999). Polyploid evolution in the European orchid genus Nigritella: evidence from DNA fingerprinting. Masters thesis, Lund University, Sweden.
Stark, C., Babik, W. & Durka, W. (2009). Fungi from the roots of the common terrestrial orchid Gymnadenia conopsea. Mycol. Res. 113: 952 – 959.
Stark, C., Michalski, S. G., Babik, W., Winterfeld, G. & Durka, W. (2011). Strong genetic differentiation between Gymnadenia conopsea and G. densiflora despite morphological similarity. Pl. Syst. Evol. 293: 213 – 226.
Strack, D., Busch, E. & Klein, E. (1989). Anthocyanin patterns in European orchids and their taxonomic and phylogenetic relevance. Phytochemistry 28: 2127 – 2139.
Sun, M., Schlüter, P. M., Gross, K. & Schiestl, F. P. (2015). Floral isolation is the major reproductive barrier between a pair of rewarding orchid sister species. J. Evol. Biol. 28: 117 – 129.
Swofford, D. L. (2003). PAUP*: Phylogenetic analysis using parsimony v4.0b10. Sinaeur, Sunderland, Massachusetts.
Tang, Y., Yukawa, T., Bateman, R. M., Jiang, H. & Peng, H. (2015). Phylogeny and classification of the East Asian Amitostigma alliance (Orchidaceae: Orchideae) based on six DNA markers. BMC Evol. Biol. 15: e96 [32 pp.]. https://doi.org/10.1186/s12862-015-0376-3.
Teppner, H. (2004). A review of new results in Nigritella (Orchidaceae). Sprawozdania z posiedzen komisji naukowych 46: 111 – 116.
Teppner, H. & Klein, E. (1985). Karyologie und Fortpflanzungsmodus von Nigritella (Orchidaceae–Orchideae), inkl. N. archiducis-joannis spec. nov. und zweier Neukombinationen. Phyton (Horn) 25: 147 – 176.
Tesitelová, T., Jersáková, J., Roy, M., Kubátová, B., Tesitel, J., Urfus, T., Travnicek, P. & Suda, J. (2013). Ploidy-specific interactions: divergence of mycorrhizal fungi between cytotypes of the Gymnadenia conopsea group (Orchidaceae). New Phytol. 199: 1022 – 1033.
Trávníček, P., Jersáková, J., Kubátová, B., Krejčíková, J., Bateman, R. M. & 22 co-authors (2012). Minority cytotypes in European populations of the Gymnadenia conopsea complex (Orchidaceae) greatly increase intraspecific and intrapopulation diversity. Ann. Bot. 110: 977 – 986.
Trávníček, P., Kubátová, B., Čurn, V., Rauchová, J., Krajniková, E., Jersaková, J. & Suda, J. (2011). Remarkable coexistence of multiple cytotypes of the Gymnadenia conopsea aggregate (the fragrant orchid): evidence from flow cytometry. Ann. Bot. 107: 77 – 87.
Tyteca, D., Ceinos, M., Gathoye, J.-L., Brys, R. & Jacquemyn, H. (2012). On the morphological, biological and genetic heterogeneity of the genus Orchis (Orhidaceae, Orchidinae). Phytotaxa 75: 19 – 32.
van der Niet, T. & Linder, H. P. (2008). Dealing with incongruence in the quest for the species tree: a case study from the orchid genus Satyrium. Molec. Phylogenet. Evol. 47: 154 – 174.
Vermeulen, P. (1947). Studies on dactylorchids. Schotanus & Jens, Utrecht.
Vermeulen, P. (1972). Übersicht zur Systematik und Taxonomie der Gattung Orchis s. str. Jber. Naturwiss. Ver. Wuppertal 25: 22 – 36.
Vermeulen, P. (1977). Orchideeën, systematisch ingedeeld. In: J. Landwehr (ed.), Wilde orchideeën van Europa, pp. 551 – 557. VBNN, Amsterdam.
Vöth, W. (2000). Gymnadenia, Nigritella und ihre Bestäuber. J. Eur. Orch. 32: 547 – 573.
Vöth, W. & Sontag, S. (2006). Die intraspezifischen Varietäten der Gymnadenia conopsea (L.) R. Br. J. Eur. Orch. 38: 581 – 624.
White, T. J., Bruns, T., Lee, S. & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: M. A. Innis, D. H. Gelfand, J. J. Sninsky & T. J. White (eds), PCR protocols: a guide to methods and applications, pp. 315 – 322. Academic Press, San Diego.
Acknowledgements
We thank the following colleagues for assistance in acquiring research materials used in this study and its predecessors: Kath Barrett, Jarle Bjerke, Mike Clark, Mihaela Nikolova, the late Tony Hughes, Nigel Johnson, Kath Fairhurst, Mikael Hedrén, David Long, Richard Manuel, Jess Stone, and Bill Temple. We also thank Petr Efimov and an anonymous reviewer for their generous reviews. RB acknowledges receipt of small but invaluable fieldwork grants from the Botanical Society of Britain and Ireland and from the Botanical Research Fund, both employed during early stages of this long-term project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Open Access
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/ licenses/by/4.0/), which permits unrestricted use, distribution and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate whether changes were made.
Accepted for publication 24 September 2018.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Bateman, R.M., Murphy, A.R.M., Hollingsworth, P.M. et al. Molecular and morphological phylogenetics of the digitate-tubered clade within subtribe Orchidinae s.s. (Orchidaceae: Orchideae). Kew Bull 73, 54 (2018). https://doi.org/10.1007/s12225-018-9782-1
Accepted:
Published:
DOI: https://doi.org/10.1007/s12225-018-9782-1