Abstract
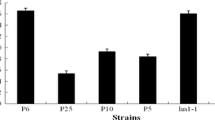
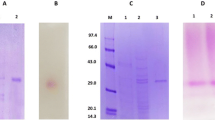
Of the 19 strains of Rhizopus delemar deposited as Rhizopus oryzae, seven of them, NBRC 4726, NBRC 4734, NBRC 4746, NBRC 4754, NBRC 4773, NBRC 4775, and NBRC 4801, completely hydrolyzed exogenous sucrose and fructooligosaccharides. The sucrose-hydrolyzing enzyme was purified from the culture filtrate of R. delemar NBRC 4754 and classified to β-fructofuranosidase, similar to that of Amylomyces rouxii CBS 438.76. Fragments including β-fructofuranosidase genes (sucA) of seven strains of R. delemar and A. rouxii CBS 438.76 were amplified and sequenced by PCR with degenerated primers synthesized on the basis of the internal amino acid sequences of purified enzymes and successive inverse PCR. Nucleotide sequences of the obtained fragments revealed that open reading frames of 1,569 bp have no intron and encode 522 amino acids. The presumed proteins contained the typical domain of the glycoside hydrolase 32 family, including β-fructofuranosidase, inulinase, levanase, and fructosyltransferases. Amino acid sequences of SucA proteins from the seven strains of R. delemar were identical and showed 90.0 % identity with those of A. rouxii CBS 438.76. A dendrogram constructed from these amino acid sequences showed that SucA proteins are more closely related to yeast β-fructofuranosidases than to other fungal enzymes.





Similar content being viewed by others
References
Abe A, Oda Y, Asano K, Sone T (2007) Rhizopus delemar is the proper name for Rhizopus oryzae fumaric-malic acid producers. Mycologia 99:714–722
Alméciga-Díaz CJ, Gutierrez AM, Bahamon I, Rodríguez A, Rodríguez MA, Sánchez OF (2011) Computational analysis of the fructosyltransferase enzymes in plants, fungi and bacteria. Gene 484:26–34
Battaglia E, Benoit I, van den Brink J, Wiebenga A, Coutinho PM, Henrissat B, de Vries RP (2011) Carbohydrate-active enzymes from the zygomycete fungus Rhizopus oryzae: a highly specialized approach to carbohydrate degradation depicted at genome level. BMC Genomics 12:38
Bernfeld P (1955) Amylases, α and β. Methods Enzymol 1:149–158
Bulut S, Elibol M, Ozer D (2004) Effect of different carbon sources on L(+)-lactic acid production by Rhizopus oryzae. Biochem Eng J 21:33–37
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37:D233–D238
Ellis JJ, Rhodes LJ, Hesseltine CW (1976) The genus Amylomyces. Mycologia 68:131–143
Ghosh B, Ray RR (2011) Current commercial perspective of Rhizopus oryzae: a review. J Appl Sci 11:2470–2486
Ghosh K, Dhar A, Samanta TB (2001) Purification and characterization of an invertase produced by Aspergillus ochraceus TS. Indian J Biochem Biophys 38:180–185
Guio F, Rodriguez MA, Alméciga-Diaz CJ, Sánchez OF (2009) Recent trends in fructooligosaccharides production. Recent Pat Food Nutr Agric 1:221–230
Guo Y, Yan QJ, Jiang ZQ, Teng C, Wang XL (2010) Efficient production of lactic acid from sucrose and corncob hydrolysate by a newly isolated Rhizopus oryzae GY18. J Ind Microbiol Biotechnol 37:1137–1143
Hesseltine CW (1983) Microbiology of oriental fermented foods. Ann Rev Microbiol 37:575–601
John RP, Anisha GS, Nampoothiri KM, Pandey A (2009) Direct lactic acid fermentation: focus on simultaneous saccharification and lactic acid production. Biotechnol Adv 27:145–152
Kito H, Abe A, Sujaya IN, Oda Y, Asano K, Sone T (2009) Molecular characterization of the relationships among Amylomyces rouxii, Rhizopus oryzae, and Rhizopus delemar. Biosci Biotechnol Biochem 73:861–864
Klein RD, Poorman RA, Favreau MA, Shea MH, Hatzenbuhler NT, Nulf SC (1989) Cloning and sequence analysis of the gene encoding invertase from the yeast Schwanniomyces occidentalis. Curr Genet 16:145–152
Lateef A, Oloke JK, Kana EBG, Oyeniyi SO, Onifade OR, Oyeleye AO, Oladosu OC (2008) Rhizopus stolonifer LAU 07: a novel source of fructosyltransferase. Chem Pap 62:635–638
Mertens JA, Skory CD (2007) Isolation and characterization of a second glucoamylase gene without a starch binding domain from Rhizopus oryzae. Enzyme Microb Technol 40:874–880
Nakamura T, Shitara A, Matsuda S, Matsuo T, Suiko M, Ohta K (1997) Production, purification and properties of an endoinulinase of Penicillium sp. TN-88 that liberates inulotriose. J Ferment Bioeng 84:313–318
Naumova ES, Serpova EV, Korshunova IV, Naumov GI (2011) Molecular polymorphism of α-galactosidase MEL genes of Saccharomyces yeasts. Microbiology 80:502–513
Oda Y, Ouchi K (1991) Construction of a sucrose-fermenting bakers' yeast incapable of hydrolysing fructooligosaccharides. Enzyme Microb Technol 13:495–498
Oda Y, Yajima Y, Kinoshita M, Ohnishi M (2003) Differences of Rhizopus oryzae strains in organic acid synthesis and fatty acid composition. Food Microbiol 20:371–375
Ohta K, Suetsugu N, Nakamura T (2002) Purification and properties of an extracellular inulinase from Rhizopus sp. strain TN-96. J Biosci Bioeng 94:78–80
Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Rehms H, Barz W (1995) Degradation of stachyose, raffinose, melibiose and sucrose by different tempe-producing Rhizopus fungi. Appl Microbiol Biotechnol 44:47–52
Roa Engel CA, Straathof AJ, Zijlmans TW, van Gulik WM, van der Wielen LA (2008) Fumaric acid production by fermentation. Appl Microbiol Biotechnol 78:379–389
Ruiz-Herrera J, González-Prieto JM, Ruiz-Medrano R (2002) Evolution and phylogenetic relationships of chitin synthases from yeasts and fungi. FEMS Yeast Res 1:247–256
Sabater-Molina M, Larque E, Torrella F, Zamora S (2009) Dietary fructooligosaccharides and potential benefits on health. J Physiol Biochem 65:315–328
Saito K, Saito A, Ohnishi M, Oda Y (2004) Genetic diversity in Rhizopus oryzae strains as revealed by the sequence of lactate dehydrogenase genes. Arch Microbiol 182:30–36
Schipper MAA (1983) A revision of the genus Rhizopus. I. The Rhizopus stronifer-group and Rhizopus oryzae. Stud Mycol 25:1–19
Schipper MAA, Stalpers JA (1984) A revision of the genus Rhizopus. II. The Rhizpopus microsporus-group. Stud Mycol 25:20–34
Severo CB, Guazzelli LS, Severo LC (2010) Chapter 7: Zygomycosis. J Bras Pneumol 36:134–141
Sharifia M, Karimi K, Taherzadeh MJ (2008) Production of ethanol by filamentous and yeast-like forms of Mucor indicus from fructose, glucose, sucrose, and molasses. J Ind Microbiol Biotechnol 35:1253–1259
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Uma C, Gomathi D, Muthulakshmi C, Gopalakrishnan VK (2010) Production, purification and characteriation of invertase by Aspergillus flavus using fruit peel waste as substrate. Adv Biol Res 4:31–36
Vially G, Marchal R, Guilbert N (2010) L(+) Lactate production from carbohydrates and lignocellulosic materials by Rhizopus oryzae UMIP 4.77. World J Microbiol Biotechnol 26:607–614
Watanabe T, Oda Y (2008) Comparison of sucrose-hydrolyzing enzymes produced by Rhizopus oryzae and Amylomyces rouxii. Biosci Biotechnol Biochem 72:3167–3173
Acknowledgments
We thank T. Watanabe, K. Sakai, and Y. Nasu for their technical assistance. This study was supported in part by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (MAFF) Rural Bio-Mass Research Project (BUM-Cm1200).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PPT 204 kb)
Rights and permissions
About this article
Cite this article
Orikasa, Y., Oda, Y. Molecular characterization of β-fructofuranosidases from Rhizopus delemar and Amylomyces rouxii . Folia Microbiol 58, 301–309 (2013). https://doi.org/10.1007/s12223-012-0211-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-012-0211-9