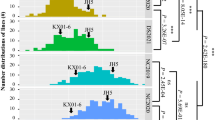
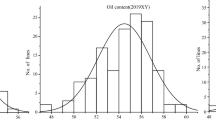
Abstract
Peanut seed oil is an important commodity worldwide and breeding efforts have been to improve both the quality and quantity of oil produced. Identifying sources of variation and elucidating the genetics of oil concentration and quality in peanut is essential to advancing the development of improved genotypes. The objective of this study was to discover QTLs for oil traits in an advanced backcross population derived from a cross between a wild-species derived amphidiploid, TxAG-6, and a cultivated genotype, Florunner. A BC1F1 population was developed for genetic mapping and an advanced backcross BC3F6 population was phenotyped in three environments and genotyped using SSR markers. Composite interval mapping results identified three genomic regions associated with oil concentration in a combined analysis. Marker PM36, associated with oil concentration and multiple fatty acids in this study, mapped directly to a HD-ZIP transcription factor in diploid Arachis genome sequences. For fatty acid concentrations, results suggested 17 QTLs identified in two or more environments, 15 of which were present across environments. Fourteen genomic regions on 13 linkage groups contained significant QTLs for more than one trait, suggesting that same genes or gene families are responsible for multiple phenotypes. QTLs and the genes identified in this study could be effective tools in marker-assisted breeding targeted at pyramiding seed oil alleles from wild-species while minimizing introgression of non-target chromatin.



Similar content being viewed by others
References
Baring MR et al. (2013) Variability of total oil content in peanut across the state of Texas. J. Crop Improv 27:125–126
Barker GC et al. (2007) Novel insights into seed fatty acid synthesis and modification pathways from genetic diversity and quantitative trait loci analysis of the Brassica C genome. Plant Physiol 144:1827–1842
Beavis, WD (1998) QTL analyses: power, precision, and accuracy. In: Paterson AH, editor. Molecular dissection of complex traits CRC Press Inc, Boca Raton pp 145–162
Belamkar V et al. (2011) A first insight into population structure and linkage disequilibrium in the US peanut minicore collection. Genetica 139:411–429
Bernacchi D et al. (1998a) Advanced backcross QTL analysis in tomato I. Identification of QTLs for traits of agronomic importance from Lycopersicon hirsutum. Theor Appl Genet 97:381–397
Bernacchi D et al. (1998b) Advanced backcross QTL analysis of tomato II. Evaluation of near-isogenic lines carrying single-donor introgressions for desirable wild QTL-alleles derived from Lycopersicon hirsutum and L. pimpinellifolium. Theor Appl Genet 97:170–180
Bernardo, R (2010) Breeding for quantitative traits in plants. Stemma Press, Woodbury, MN
Bertioli DJ et al. (2016) The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. doi:10.1038/ng.3517
Broman KW, Wu H, Sen Ś, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Brothers AN et al. (2013) Genetic architecture of floral traits in Iris hexagona and Iris fulva. J Hered 104:853–861
Burow MD, Simpson CE, Starr JL, Paterson AH (2001) Transmission of genetics of chromatin from a synthetic amphidiploid to cultivated peanut (Arachis hypogaea L.): broadening the gene pool of a monophyletic polyploid species. Genetics 159:823–837
Burow, M.D et al (2013) Marker-Assisted selection for biotic stress resistance in peanut. In: Varshney RK, and Tuberosa R, editors. Translational genomics for crop breeding: biotic stress, Vol I. John Wiley & Sons Ltd, Chichester. doi:10.1002/9781118728475.ch8
Burow, MD et al (2014a). Identification of additional FAD2 genes plus DGAT genes in peanut and mapping QTLs for fatty acid composition in peanut. 46th Annual Meeting of the American Peanut Research and Education Society, San Antonio, Texas. 10 July 2014. Paper 72
Burow MD et al. (2014b) Introgression of homeologous quantitative trait loci (QTLs) for resistance to the root-knot nematode [Meloidogyne arenaria (Neal) Chitwood] in an advanced backcross-QTL population of peanut (Arachis hypogaea L.). Mol. Breeding 34:393–406
Chai G et al. (2010) Brassica GLABRA2 genes: analysis of function related to seed oil content and development of functional markers. Theor. Appl. Genet 120:1597–1610
Chopra R et al. (2015) Next-generation transcriptome sequencing, SNP discovery and validation in four market classes of peanut, Arachis hypogaea L. Mol. Genetics and. Genomics 290:1169–1180
Davis JP, Geller D, Faircloth WH, Sanders TH (2009) Comparisons of biodiesel produced from unrefined oils of different peanut cultivars. J Am Oil Chem Soc 86:252–236
Eskandari M, Cober ER, Rajcan I (2013a) Genetic control of soybean oil I: QTL and genes associated with seed oil accumulation in RIL populations derived from crossing moderately high-oil parents. Theor Appl Genet 126:483–495
Eskandari M, Cober ER, Rajcan I (2013b) Using the candidate gene approach for detecting genes underlying seed oil concentration and yield in soybean. Theor Appl Genet 126(7):1839–1850
Fulton TM et al. (2000) Advanced backcross QTL analysis of a Lycopersicon esculentum x Lycopersicon parviflorum cross. Theor Appl Genet 100:1025–1042
Goh T et al. (2007) VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell 19:3504–3515
Gomez, SM et al (2008) Towards and integrated SSR/RFLP map of tetraploid peanut. Third International Conference of the Peanut Research Community, ICRASAT, Hyderabad, Andhra Pradesh, India. 4–8 Nov. 2008. Paper 21
Gomez SM, Narayana M, Schubert AM, Ayers JL, Baring MR, Burow MD (2009) Identification of QTLs for pod and kernel traits in cultivated peanut by bulked segregant analysis. Electr J Biotech 12(2):1–10
Graef G et al. (2009) A high-oleic and low-palmitic-acid soybean: agronomic performance and evaluation as a feedstock for biodiesel. Plant Biotechnology J. 7:411–421
Isleib TG, Pattee HE, Giesbrecht FG (2004) Oil, sugar, and starch characteristics in peanut breeding lines selected for low and high oil content and their combining ability. J Agric Food Chem 52:3165–3168
Isleib TG, Wilson RF, Novitzky WP (2006) Partial dominance, pleiotropism, and epistasis in the inheritance of the high-oleate trait in peanut. Crop Sci 46:1331–1335
Jing Z et al. (2010) QTL analysis of yield-related traits using an advanced backcross population derived from common wild rice (Oryza rufipogon L). Mol. Plant Breed 1:2–10
Jung S, Powell G, Moore K, Abbott A (2000) The high oleate trait in the cultivated peanut [Arachis hypogaea L.] II. Molecular basis and genetics of the trait. Mol Gen Genet 263:806–811
Jungman, B (2000) The effect of fatty acid profiles on peanut seed germination at low soil temperatures. M.S. thesis., Texas Tech Univ., Lubbock
Lardizabal K et al. (2008) Expression of Umbelopsis ramanniana DGAT2A in seed increases oil in soybean. Plant Physiol 148:89–96
Larson SR, Mayland HF (2007) Comparative mapping of fiber, protein, and mineral content QTLs in two interspecific Leymus wild rye full-sib families. Mol Breeding 20:331–347
Liu YF et al. (2014) Soybean GmMYB73 promotes lipid accumulation in transgenic plants. BMC Plant Biol 14:–73
López Y et al. (2000) Isolation and characterization of the Δ12-fatty acid desaturase in peanut (Arachis hypogaea L.) and search for polymorphisms for the high oleate trait in Spanish market-type lines. Theor Appl Genet 101:1131–1138
López Y, Smith OD, Senseman SA, Rooney WL (2001) Genetic factors influencing high O/L acid content in Spanish market-type peanut cultivars. Crop Sci 41:51–56
Lung SC, Weselake RJ (2006) Diacylglycerol acyltransferase: a key mediator of plant triacylglycerol synthesis. Lipids 41:1073–1088
Mergeai G (2006) Cotton improvement through interspecific hybridization. Cahiers. Agri 15:135–143
Moore KM, Knauft DA (1989) The inheritance of high oleic acid in peanut. Jour Hered. 80:252–253
Norden AJ, Lipscomb RW, Carver WA (1969) Registration of ‘Florunner’ peanuts. Crop Sci 9:850
O’Byrne DJ, Knauft DA, Shireman RB (1997) Low fat monounsaturated rich diets containing high-oleic peanuts improve serum lipoprotein profiles. Lipids 32:687–695
Pandey MS et al. (2014) Identification of QTLs associated with oil content and mapping FAD2 genes and their relative contribution to oil quality in peanut (Arachis hypogaea L. BMC Genet 15:–133
Percival AE, Wendel JF, Stewert JM (1999) Taxonomy and germplasm resources. In: Smith CW, Cothren JT (eds) Cotton: Origin, history, technology, and production. John Wiley & Sons, New York, pp. 33–63
Ramos MJ et al. (2009) Influence of fatty acid composition of raw materials on biodiesel properties. Bioresource Tech 100:261–268
Ros E, Mataix J (2006) Fatty acid composition of nuts – implications for cardiovascular health. British. J Nutr 96:S29–S35
Saha S, Enugutti B, Rajakumari S, Rajasekharan R (2006) Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferace. Plant Physiol 141:1533–1543
Sarvamangala C, Gowda MVC, Varshney RK (2011) Identification of quantitative trait loci for protein content, oil content and oil quality in groundnut (Arachis hypogaea L. Field Crops Res 122:49–59
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nature Biotech 18:233–234
Selvaraj MG et al. (2009) Identification of QTLs for pod and kernel traits in cultivated peanut by bulked segregant analysis. Electronic J. Of. Biotech 12. doi:10.2225/vol12
Shen B, Sinkevicius KW, Selinger DA, Tarczynski MC (2006) The homeobox gene GLABRA2 affects seed oil content in Arabidopsis. Plant Mol Biol 60:377–387
Shi L et al. (2012) Arabidopsis glabra2 mutant seeds deficient in mucilage biosynthesis produce more oil. Plant J 69:37–46
Simpson CE (1991) Pathways for introgression of pest resistance into Arachis hypogaea L. Peanut Sci. 18:22–26
Simpson CE, Starr JL (2001) Registration of ‘COAN’ peanut. Crop Sci 41:918
Simpson CE et al. (1993) Registration of ‘TxAG-6’ and ‘TxAG-7’ peanut germplasm. Crop Sci 33:–1418
Tanksley SD, Nelson JC (1996) Advanced backcross QTL analysis: a method for the transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92:191–203
Tian F et al. (2006) Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (O. sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor. Appl. Genet 112:570–580
Vassiliou EK et al. (2009) Oleic acid and peanut oil high in oleic acid reverse the inhibitory effect of insulin production of the inflammatory cytokine TNF-alpha both in vitro and in vivo systems. Lipids Health Dis 8:–25
Wang ML et al. (2015) Genetic mapping of QTLs controlling fatty acids provided insights into genetic control of fatty acid synthesis pathway in peanut (Arachis hypogaea L.). Plos One. doi:10.1371/journal.pone.0119454
Wilson JN et al. (2013a) Generation means analysis of fatty acid composition in peanut. J. Crop Improv 27:430–443
Wilson JN et al. (2013b) Diallel analysis of oil production components in peanut (Arachis hypogaea L.). Int J Agro. doi:10.1155/2013/975701
Wilson JN et al. (2013c) Generation means analysis of oil concentration in peanut. J. Crop Improv 27:85–95
Xiang L, Etxeberria E, Van den Ende W (2013) Vacuolar protein sorting mechanisms in plants. FEBS J 280:979–993. doi:10.1111/febs.12092
Zheng P et al. (2008) A phenylalanine in DGAT is key determinant of oil content and composition in maize. Nat Genet 40:367–372
Acknowledgments
This work was funded by USDA/NIFA award TEX08835 to MDB, National Peanut Board award #329/TX-99 to MRB, MDB, and CES, and through support provided by the Office of Agriculture, Research and Policy, Bureau of Food Security, U.S. Agency for International Development, under the terms of Award No. AID-ECG-A-00-07-0001 to The University of Georgia as management entity for the U.S. Feed the Future Innovation Lab on Peanut Productivity and Mycotoxin Control. The opinions expressed herein are those of the author(s) and do not necessarily reflect the views of the U.S. Agency for International Development.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Ray Ming
Key message: QTLs, some with large phenotypic effects, were observed for oil and fatty acid concentrations. Marker PM36 mapped directly to a HD-ZIP transcription factor in diploid Arachis genome sequences.
Electronic supplementary material
Table S1
(DOCX 18 kb)
Rights and permissions
About this article
Cite this article
Wilson, J.N., Chopra, R., Baring, M.R. et al. Advanced Backcross Quantitative Trait Loci (QTL) Analysis of Oil Concentration and Oil Quality Traits in Peanut (Arachis hypogaea L.). Tropical Plant Biol. 10, 1–17 (2017). https://doi.org/10.1007/s12042-016-9180-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-016-9180-5