Abstract
In the present study, the genetic variability of the EG95 protein-coding gene in several animal and human isolates of Echinococcus granulosus was investigated. A total of 24 isolates collected from cattle, buffalo, sheep, goat, dog and man were amplified by Eg95-coding gene-specific primers. From the generated sequence information, a conceptual amino acid sequence was deduced. Phylogenetically, the Eg95 coding gene belongs to the Eg95-1/Eg95-2/Eg95-3/Eg95-4 cluster. Further confirmation on the maximum composite likelihood analysis revealed that the overall transition/transversion bias was 2.913. This finding indicated that there is bias towards transitional and transversional substitution. Using artificial neural networks, a B-cell epitope was predicted on primary sequence information. Stretches of amino acid residues varied between animal and human isolates when hydrophobicity was considered. Flexibility also varied between larval and adult stages of the organism. This observation is important to develop vaccines. However, cytotoxic T-lymphocyte epitopes on primary sequence data remained constant in all isolates. In this study, agretope identification started with hydrophobic amino acids. Amino acids with the same physico-chemical properties were present in the middle. The conformational propensity of the Eg95-coding gene of 156 amino acid residues had \(\upalpha \)-turns and \(\upbeta \)-turns, and \(\upalpha \)-amphipathic regions up to 129, 138–156 and 151–155 residues, respectively. The results indicated potential T-cell antigenic sites. The overall Tajima’s D value was negative (−2.404165), indicative of negative selection pressure.
Similar content being viewed by others
Introduction
Hydatidosis or cystic echinococcosis (CE) caused by the larval stage of the tapeworm Echinococcus granulosus is a classic example of cyclozoonosis (Eckert and Thompson 1997). The parasite is distributed worldwide (Nunnari et al. 2012) and causes significant economic losses to the meat industry, mainly due to condemnation of edible offal namely liver and lungs of food animals (Torgerson and Heath 2003). Besides, the disease has a potential public health impact that is recognized by the World Health Organization (WHO 2003). Echinococcosis in man is often expensive and difficult to treat. Sometimes treatment of the disease requires either extensive surgery or prolonged chemotherapy or the simultaneous use of both (WHO 2018). In Asia and Africa, around 50 million people are at risk of developing the disease (Hemphill and Kern 2008). Office International des Epizooties has recognized CE as a multispecies disease since this species has low intermediate host specificity and infects domestic and wild ungulates, and humans (Thompson and McManus 2001; Gauci et al. 2002). Intermediate hosts get the infection after the ingestion of parasite eggs excreted by the canid definitive host (Alvarez-Rojas et al. 2012). Within the intermediate host, the unilocular cyst develops mainly in the lung and liver (Torgerson and Heath 2003). The cyst contains an inner germinal membrane that is externally supported by an acellular laminated layer. By clonal expansion of the germinal membrane, protoscoleces develop (Thompson et al. 1995).
Within the host, the helminth parasites survive for a long time. This is possible due to the developed and altered strategies to feed and reproduce after evading host immune attack. The evasion mechanism is mediated by proteins that are collectively known as secreted and membrane bound (S/M) proteins. S/M proteins induce apoptosis of the dendritic cells (Nono et al. 2012) and produce an immunosuppressive environment in the infection phase (Huang et al. 2016). Since these proteins are expressed before the host immune system, S/M components are candidates for ameliorated diagnosis tests as well as a target for new drugs and immunoprophylactic agents (Rosenzvit et al. 2006). A total of 12 S/M proteins have been reported as immunodiagnostic and immunoprophylactic agents (Rosenzvit et al. 2006). Of these, Eg95 is a candidate antigen used for immunization of animals. This biomolecule is expressed in the oncosphere and is used for immunization against the larval stage of the disease. Earlier reports suggest that there was a 62% decrease in the prevalence of infection in sheep, which occurs due to the development of the larval stage of the parasite (Lightowlers and Heath 2004; Larrieu et al. 2015). This antigen is expressed by oncospheres, protoscoleces and immature and mature adult worms (Zhang et al. 2003). A vaccine has been developed based on a recombinant antigen of EG95 and it has been shown to be effective in the prevention of the disease in different intermediate hosts (Lightowlers et al. 1996, 1999). A perusal of the available literature suggests that EG95 is a multigene family and comprises seven isoforms (EG95-1 to EG95-7). All the isoforms except EG95-7 comprise three exons that are separated by two introns (Chow et al. 2001). Exon 1 encodes a signal peptide and exons 2 and 3 encode mature peptides (Haag et al. 2009).
There is accumulated evidence to show that antigens encoded by a multigene family show a high degree of polymorphism (Arend et al. 2004; Kamenetzky et al. 2005). This occurs due to natural selection pressure which in turn leads to the diversification of antigen-coding genes. There are two models of natural selection: purifying selection (negative selection) and directional selection (positive selection) (Haag et al. 2009). The selection pressure of an antigen-coding gene is exclusively dependent on synonymous and nonsynonymous substitution which occurs due to a change in the nucleotide sequence (Pan et al. 2010a). An antigen-coding gene undergoing positive selection pressure exhibits an excess of nonsynonymous substitution. This type of substitution indicates the heterogeneous selection pressure exerted by the host (Haag et al. 2009). On the other hand, large deletions are mainly found within introns 1 and 2 of this antigen-coding gene which is a footprint of gene conservation (Haag et al. 2009). Therefore, the use of a vaccine based on single recombinant antigens may select for antigenically variant parasites that are ineffective as vaccines (Boubaker et al. 2014). Hence, the practical use of the developed EG95 vaccine may be affected by the antigenic variability of the EG95 protein expressed in the different intermediate hosts. This problem can be overcome by monitoring antigenic variants among parasite populations and incur necessary modification for appropriate vaccine development (Lightowlers et al. 2003). Moreover, extensive isolate variations have been observed in the parasite with 10 designated genotypes (G1–G10) that may impact the epidemiology, pathology and control of CE (Spotin et al. 2017).
Allelic polymorphisms of the Eg95 gene may impact the efficacy of the vaccine based on Eg95 (Gauci et al. 2018). Therefore, understanding the polymorphism of the EG95 genes in different E. granulosus isolates is very important. Moreover, it has been observed that EG95 epitopes are conformational (Woollard et al. 2000), indicating that a few amino acid substitutions may affect protection dramatically (Haag et al. 2009). A crucial step to design a peptide vaccine involves the identification of T cell, B cell and cytotoxic T-lymphocyte (CTL) epitopes. The experimental scanning of the epitopes requires the synthesis of overlapping peptides that span the entire sequence of a protein antigen. But this approach is costly and labour intensive. The alternative approach is to find out the epitopes by in silico analysis (Bhasin and Raghava 2004). In the present study, we investigated the genetic variability of the EG95 protein-coding gene in several animal and human isolates of E. granulosus. The antigenic variability of the deduced EG95 proteins was also assessed, and B and T cell epitopes were predicted to have huge implications for stain-specific vaccine development.
Materials and methods
Chemicals, reagents, media and buffers
Molecular biology and analytical grade chemicals were procured from Fermentas, Hi-Media, Amresco, Progema, SRL, CISCO, Merck and Sigma. Media and buffers used in this study were purchased from Hi-Media, SRL and Difco.
Collection of parasite samples
Cyst samples from animals were collected from local abattoirs situated at Kolkata, West Bengal. Adult parasites were collected from experimentally infected pups. Human cyst samples were collected from RG Kar Medical College Hospital, Kolkata, West Bengal, India. The collected samples were transported to the laboratory by maintaining an appropriate cold chain. Parasitic samples from 21 animals and three human patients were collected (table 1).
Isolation of genomic DNA
Genomic DNA from fresh protoscoleces of individual hydatid cysts and from adult worms was extracted using a commercial kit (Q-BIOgene, Carlsbad, USA) as per the protocol of the manufacture, as previously described (Pan et al. 2010b). Following the phenol–chloroform extraction, genomic DNA was precipitated with isopropanol and dissolved in Tris-EDTA buffer (pH 8.0) and kept at \(-80^{\circ }\hbox {C}\) until further use.
Identification of the genotype of isolates of E. granulosus
The identification of the genotype was performed using a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method as described previously (Pan et al. 2009). In brief, a 770-bp fragment was amplified by PCR and the amplified fragment was digested with Alu1 and Casp61 restriction enzymes. Genotypes were identified by restriction profiles of the fragment (Pan et al. 2009).
Amplification of the EG95 antigen-coding region
The EG95-coding region was amplified by PCR using primers described by Haag et al. (2009). The PCR was carried out in a total volume of \(25\, \mu \hbox {L}\) containing DNA (100 ng), \(200\, \mu \hbox {M}\) of deoxyribonucleotide triphosphates, \(10\times \) PCR buffer, \(2.5\hbox { mM MgCl}_{2}\), 10 pmol each of forward and reverse primers and 1 unit of Taq DNA polymerase. The PCR was carried out in an initial denaturation (\(94^{\circ }\hbox {C}\)) for 5 min followed by 37 cycles. For the first two cycles, the annealing temperature was \(60^{\circ }\hbox {C}\), followed by \(55^{\circ }\hbox {C}\) for the next 10 cycles with decrement of \(1^{\circ }\hbox {C}\) after each cycle. For the last 25 cycles, the annealing temperature was \(45^{\circ }\hbox {C}\). For all the cycles, 30 s each was given for denaturation and annealing, and 1 min was given for extension. The confirmation of the amplicon was performed by agarose gel electrophoresis (figure 1).
Cloning of the PCR product and sequencing
Amplicons were cloned into the pTZ57R/T vector using the InsTAclone PCR cloning kit (Fermentas, Burlington, USA) following the manufacturer’s protocol. The ligation product was transformed into E. coli DH5\(\upalpha \) competent cells. The recombinant clones were identified by blue/white selection and the positive white colonies were analysed for the confirmation of correct insert and orientation in the purified plasmids by the colony PCR and by double digestion with EcoR1 and BamH1 (figure 1). Then, the purified plasmids were sequenced with standard primers (M13F and M13R) using a chain termination method.
Prediction of promiscuous major histocompatibility complex (MHC)-II binding site, B cell epitope, CTL epitope, antigenic index and conformational propensity
The prediction of the promiscuous MHC-II binding site was evaluated as described earlier by Singh and Raghava (2001). The B-cell epitope was predicted using online software developed by Saha and Raghava (2006). The CTL epitope was predicted as described by Bhasin and Raghava (2004). An antigenic index and conformational propensity were deduced by Lasergene 8.0 (DNA Star, USA) and a PSIPRED secondary structure prediction method (Jones 1999). Tajima’s neutrality test and transition/transversion bias were calculated using MEGA v5.1 (Tamura et al. 2007).
Phylogenetic analysis
Multiple sequence alignment of the deduced amino acid sequences was performed using Clustal W. Phylogenetic analysis was performed by the distance-based neighbour-joining method using MEGA 5.1 (CEMI, Tempe, USA). Tree evaluation was performed by bootstrapping for 1000 replicates to test the accuracy of the phylogenetic tree. Sequence information was compared with the available sequence information of EG 95-1 (AAL35393), Eg95-2 (AAG40127), Eg95-3 (AAG40128), Eg95-4 (AAG40124), Eg95-5 (AAG40125) and Eg95-6 (AAG40123).
Results
Genotype of isolates of E. granulosus
The genotype of eight buffalo, two cattle, five sheep, five goat, three human isolates and one adult worm was performed through PCR-RFLP. It was found that all isolates belonged to the common sheep strain (G1) except one buffalo isolate that was a cattle strain (G5) (table 1).
Pairwise amino acid sequence variation
The nucleotide sequences of the EG95-coding region of 24 isolates used in the study were generated and the sequences were deposited in a public database (NCBI with accession numbers HM345583–HM345607). The conceptual amino acid sequences were deduced using universal genetic code. The multiple sequence alignment based on the deduced-amino acid sequences along with their conserved and variable regions of the 24 isolates used in the study and the EG95 of six isoforms is presented in figure 2. On the basis of the amino acid sequence variation, one goat (G8) and two sheep (S3 and S4) isolates were indistinguishable from each other. Similarly, both goat isolates (G4 and G12) and three human isolates (H1, H2 and H3) were indistinguishable from each other. Similarity of the amino acid sequence was also observed between two goat isolates (G1 and G3) and one buffalo isolate (B1). Sequence variation between 24 isolates varied from 1.21 to 11.56% (table 2).

Phylogenetic analysis
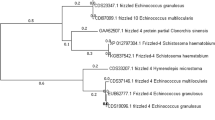
From the phylogenetic analysis, it was revealed that four isoforms of the EG95 gene described in the centralized repositories of the public database (Eg95-1, EG95-2, Eg95-3 and Eg95-4) belonged to one cluster. On the other hand, EG95-5 and EG95-6 belonged to another cluster. The isolates characterized in the present study were homologous to four isoforms (Eg95-1, Eg95-2, Eg95-3 and Eg95-4) of the Eg95-coding gene (figure 3).
The probability of substitution was estimated by using the maximum composite likelihood estimate. Transitional and transversional substitution varied from 13.08 to 27.26 and 2.68 to 3.33, respectively. The overall transition/transversion bias (R) was 2.913. The patterns of nucleotide substitution for both purine and pyrimidine bases are shown in table 3.
Variation with respect to stretches of \(\upalpha \)-helix, \(\upbeta \)-sheet, \(\upalpha \)-amphipathic and \(\upbeta \)-amphipathic was not observed between animal and human isolates. Positive values of the antigenic index were seen up to 131–137 amino acid residues in both animal and human isolates (table 4).
Physico-chemical properties of the amino acids
Physico-chemical properties of the deduced-amino acid sequence of the 24 isolates are presented in table 5. The positive GRAVY value indicates that the protein is polar in nature.
Prediction of B-cell and CTL epitopes
The prediction of the B-cell epitope was done on the basis of seven physico-chemical properties. Stretches of amino acid sequences varied between animal and human isolates when hydrophobicity was considered. Stretches of amino acid sequences varied between the larval stage and adult stage of the parasite when flexibility was considered.
Phylogenetic analysis on the amino acid sequence of the Eg95 coding gene of E. granulosus by the unweighted pair group method. Numbers at nodes represent percentage occurrence of clades in 1000 bootstrap replications (B, buffalo isolate; C, cattle isolate; S, sheep isolate; G, goat isolate; Adult, adult worm; H, human isolate) and conformational propensity of the individual isolate.
Further, stretches of amino acid sequences varied between the larval stage of animal and human isolates, as well as the adult stage of canine isolates when accessibility was taken into consideration. Only one stretch (LRNHFNLT) of the amino acid sequence was seen within the sequence information of the amino acid sequence of the adult stage of E. granulosus when turns within the sequence were assessed. Stretches of amino acid sequences varied between animal and human isolates when the exposed surface was considered. However, stretches of amino acid sequences did not vary between all isolates when polarity and antigenic propensity were considered (table 6).
The prediction of the CTL epitope revealed three stretches of CTL epitopes within 164 amino acid residues which started at 48, 92 and 142 positions. Detailed information on CTL epitopes is depicted in table 7.
Prediction of the promiscuous MHC-II binding site
The larval stage isolated from animals exhibited more than 50% binding propensity with 17 MHC-II alleles. The larval stage of human isolates showed more than 50% binding propensity with 13 MHC-II alleles. The adult stage of E. granulosus presented more than 50% binding propensity with 18 MHC-II alleles (table 8).
Selection pressure of the EG95-coding gene
Twenty-four isolates considered in the study had 326 segregating sites. By sliding window analysis, the sites of nonsynonymous and synonymous valley were analysed. The highest Tajima’s D value (\(-1.8923\)) was observed within 1–101 nucleotide bases. The lowest Tajima’s D value was observed within 302–401 nucleotide stretches (table 9, figure 4). The overall D value of 471 bases was − 2.404165. The value was statistically significant (\(P < 0.001\)).
Discussion
E. granulosus has a worldwide distribution and is an important zoonotic parasite. Although domestic livestock are its most important host, numerous herbivorous wild life species may also act as intermediate hosts (Jenkins and Macpherson 2003). Of the 10 reported genotypes (G1–G10), common sheep strain (G1) is promiscuous in its range of suitable intermediate host species (Barnes et al. 2009). This strain is distributed in parts of South America, southern and eastern Europe, northern and eastern Africa and parts of Asia and the Australian region (Eckert and Thompson 1997). Most organisms isolated from human patients have been confirmed as belonging to the common sheep strain (Arbabi et al. 2017). In the present study, of the 24 isolates of E. granulosus (both larvae and adults), 23 belonged to the common sheep strain (table 1). Therefore, considering the zoonotic significance and importance of the strain, control of the transmission of this organism is warranted.
Control of the disease is possible by preventing the infection of the definitive host or by immunization of the intermediate hosts. Immunization against the parasite is possible after understanding the protective immune response that allows for the identification of antigens eliciting the protective immune response (Craig et al. 2007). Immunization against the parasite may be performed by using S/M proteins that are expressed by the parasite (Rosenzvit et al. 2006). Previous vaccination trials indicated that EG95 induced 95% protection against hydatid infection in an experimental trial (Lightowlers et al. 1996). Later on, the vaccine was proved to be effective in Australia, New Zealand, Argentina and China (Lightowlers et al. 1999; Larrieu et al. 2015). The vaccine has also been attempted to be used in wild animals with 96–100% protective efficacy (Barnes et al. 2009).
In the past, based on preliminary reports, it was postulated that Eg95 has broad applicability as a vaccine (Lightowlers et al. 1999). This conclusion was arrived since there was no available information on the genetic variability of the gene encoding Eg95 antigen, but with the increase noesis in molecular biology, it has been worked out that the Eg95-encoding gene is a member of a multigene family (Zhang et al. 2003). DNA sequence analysis of cloned genomic DNA indicated that the Eg95 gene family consists of at least seven members, one of which (Eg95-7) is a pseudogene (Chow et al. 2001). Therefore, the use of a vaccine based on a single recombinant antigen (Eg95) may select for antigenitically variant parasites that are insusceptible to the vaccine. This problem may be ducked by monitoring the parasite population for antigenic variants and modifying the vaccine appropriately (Lightowlers et al. 2003).
As far as the phylogenetic relationship is concerned, Eg95-1 and Eg95-2, and Eg95-3 and Eg95-4, belong to one cluster. In contrast, Eg-95-5 and Eg-95-6 belong to another cluster and have 76–77% similarity with Eg95-1 (Chow et al. 2001). In the present study, from phylogenetic analysis (figure 3) and pairwise amino acid sequence variation (figure 2), it can be concluded that all isolates belonged to the Eg95-1/Eg95-2/Eg-95-3/Eg95-4 cluster. Therefore, from the present findings, it may be concluded that both the strains considered during the study (G1 and G5) can be used without any chance of vaccine resistance due to the absence of antigenic flexibility which obviously excludes the chances of common serotype replacement (Kennedy and Read 2017). This finding is helpful because proteins expressed by Eg95-1 and other members of the cluster are host protective while the protective efficacy of Eg95-5 and Eg95-6 is yet to be assessed (Lightowlers et al. 2003).
Due to nucleotide substitution, the antigenicity and efficacy of the EG95 vaccine may vary in G6/G7 genotypes.
The identification of the B-cell epitope has three different implications, namely development of antibody therapeutics (Regenmortel 2006), peptide-based vaccines (Dudek et al. 2010) and immunodiagnostics (Leinikki et al. 1993). It has been clearly described in the past that over 70% of the discontinuous B-cell epitopes are composed of 1–6 amino acids. In the present investigation, short stretches of discontinuous B-cell epitopes have been predicted (table 6). The correlations between B-cell epitope localization to its physico-chemical properties (table 6) are important observations for the development of the EG95 vaccine, which has been perceived earlier as well (Pellequer et al. 1991). For B-cell epitope formation, hydrophilicity and antigenic propensity are primary factors although other inter-related factors like flexibility cannot be ignored (Li et al. 2013). Therefore, the difference in the predicted B-cell epitope among larval stages of animal and human isolates and canine isolate (table 6) is a very important observation for designing the EG95 vaccine when the vaccine has to be developed from two different stages of the parasite. This information seems to be the pioneer observation on the development of the EG95 vaccine and has not been reported earlier.
The identification of the CTL epitope is crucial to understanding the roles of T-cell activation and designing of synthetic vaccines (Brunak and Buus 2000). In the present study, the predicted CTL epitopes of the Eg95-coding gene have remained constant in terms of distribution in all the isolates (table 7).
The T cell is activated after the formation of the trimolecular complex between its antigenic binding receptor, an MHC molecule and an antigenic peptide. Therefore, antigens recognized by the T cell have two distinct interaction sites. One is the epitope which interacts with the T-cell receptor and other is the agretope. The agretope interacts with the MHC molecule. Peptides that bind to class II MHC molecules contain a sequence of 7–10 amino acids. This sequence provides a major contact point with the MHC-II molecule (Goldsby et al. 2000). In the present study, stretches of the agretope varied from 8–10 amino acid residues (table 8). Stretches of the agretope contained aromatic or hydrophobic residues at the amino terminus and three additional hydrophobic residues in the middle (Goldsby et al. 2000). In the present study, the agretope identified on the Eg95-coding gene started with hydrophobic amino acids (either with phenylalanine or isoleucine). The amino acids with the same physico-chemical properties (hydrophobic) were present in the middle of the agretope. The agretope with MHC-II binding propensity (more than 50%) indicated a potential interaction site for the formation of the trimolecular complex.
Two antigenic properties contribute to antigen processing, presentation and recognition. The properties are amphipathicity and \(\upalpha \)-helicity. Most helper T-cell antigenic sites are amphipathic and have \(\upalpha \)-helices. Under certain circumstances, T-cell antigenic sites exhibit a \(\upbeta \)-sheet (Spouge et al. 1987). The conformational propensity of the Eg95-coding gene of 156 amino acid residues revealed \(\upalpha \)- turns and \(\upbeta \)-turns and \(\upalpha \)-amphipathic regions up to 129, 138–156 and 151–155 residues, respectively, in both animal and human isolates. This result indicated the potential T-cell antigenic site on the Eg95-coding gene. Positive values of the antigenic index were seen up to 137 residues. This finding indicated the presence of the immunodominant helper T-lymphocyte antigenic site.
Tajima’s D test value (− 2.404165) has indicated negative/purifying selection pressure that varied within stretches of the EG95 antigen. This result indicated that there was an excess of low-frequency polymorphisms within 1–101 nucleotide bases compared to other stretches of nucleotide sequences (table 9). Earlier literature suggests that there are two types of natural selection pressures in universal biological evolution: positive (Darwinian) and negative/purifying. The latter selection pressure is responsible for detrimental pseudogenization (Zhang 2008). Therefore, negative selection pressure of the EG95 antigen depicts that the biomolecule can be expressed and used for the development of vaccines without pseudogenization.
In summary, this study dealt with the genetic variability of the EG95 protein-coding gene in 24 animal and human isolates of E. granulosus. All isolates belonged to one cluster and were closely similar to four isoforms of the EG95 antigen. Epitope mapping analysis revealed that physico-chemical properties of the amino acids present on the B-cell linear epitope varied between human and animal isolates as well as between larval and adult stages of the parasites. The agretope present on the Eg95-coding gene showed the presence of hydrophilic amino acids and conformational propensity deduced on the basis of the \(\upalpha \)-helix and \(\upbeta \)-sheet indicated the potential T-cell antigenic receptor site which helps in the presentation, processing and recognition of antigens.
References
Alvarez-Rojas C. A., Gauci C. G., Nolan M. J., Harandi M. F. and Lightowlers M. W. 2012 Characterization of the Eg95 gene family in the G6 genotype of Echinococcus granulosus. Mol. Biochem. Parasitol. 183, 115–121.
Arbabi M., Pirestani M., Delavari M., Hooshyar H., Abdoli A. and Sarvi S. 2017 Molecular and morphological characterizations of Echinococcus granulosus from human and animal isolates in Kashan, Markazi Province, Iran. Iran J. Parasitol. 12, 177–187.
Arend A. C., Zaha A., Ayala F. J. and Haag K. L. 2004 The Echinococcus granulosus antigen B shows a high degree of genetic variability. Exp. Parasitol. 108, 76–80.
Barnes T. S., Hinds L. A., Jenkins D. J., Coleman G. T., Colebrook A. L., Kyngdon C. T. et al. 2009 Efficacy of the EG95 hydatid vaccine in a macropodid host, the tammar wallaby. Parasitology 136, 461–468.
Bhasin M. and Raghava G. P. 2004 Prediction of CTL epitopes using QM, SVM and ANN techniques. Vaccine 22, 3195–3204.
Boubaker G., Gottstein B., Hemphill A., Babba H. and Spiliotis M. 2014 Echinococcus P29 antigen: molecular characterization and implication on post-surgery follow-up of CE patients infected with different species of the Echinococcus granulosus complex. PLoS One 9, e98357.
Brunak S. and Buus S. 2000 Identifying cytotoxic T cell epitopes from genomic and proteomic informat ion: ‘The human MHC project’. Rev. Immunogenet. 2, 477.
Chow C., Gauci C. G., Cowman A. F. and Lightowlers M. W. 2001 A gene family expressing a host-protective antigen of Echinococcus granulosus. Mol. Biochem. Parasitol. 118, 83–88.
Craig P. S., McManus D. P., Lightowlers M. W., Chabalgoity J. A., Garcia H. H., Gavidia C. M. et al. 2007 Prevention and control of cystic echinococcosis. Lancet Infect. Dis. 7, 385–394.
Dudek N. L., Perlmutter P., Aguilar M. I., Croft N. P. and Purcell A. W. 2010 Epitope discovery and their use in peptide based vaccines. Curr. Pharm. Des. 16, 3149–3157.
Eckert J. and Thompson R. C. A. 1997 Intraspecific variation of Echinococcus granulosus and related species with emphasis on their infectivity to humans. Acta Trop. 64, 19–34.
Gauci C., Merli M., Muller V., Chow C., Yagi K., Mackenstedt U. et al. 2002 Molecular cloning of a vaccine antigen against infection with the larval stage of Echinococcus multilocularis. Infect. Immun. 70, 3969–3972.
Gauci C. G., Alvarez-Rojas C. A., Chow C. and Lightowlers M. W. 2018 Limitations of the Echinococcus granulosus genome sequence assemblies for analysis of the gene family encoding the EG95 vaccine antigen. Parasitology 145, 807–813.
Goldsby R. A., Kindt T. J. and Osborne B. A. 2000 Kuby immunology, 4th edition, pp. 63–81, 173–198. W. H. Freeman, New York.
Haag K. L., Gottstein B. and Ayala F. J. 2009 The EG95 antigen of Echinococcus spp. contains positively selected amino acids, which may influence host specificity and vaccine efficiency. PLoS One 4, e5362.
Hemphill A. and Kern P. 2008 Special issue: experimental studies in echinococcosis. Exp. Parasitol. 119, 437–438.
Huang F., Dang Z., Suzuki Y., Horiuchi T., Yagi K., Kouguchi H. et al. 2016 Analysis on gene expression profile in oncospheres and early stage metacestodes from Echinococcus multilocularis. PLoS Negl. Trop. Dis. 10, e0004634.
Jenkins D. J. and Macpherson C. N. 2003 Transmission ecology of Echinococcus in wild-life in Australia and Africa. Parasitology 127, S63–S72.
Jones D. T. 1999 Protein secondary structure prediction based on position specific scoring matrices. J. Mol. Biol. 292, 195–202.
Kamenetzky L., Muzulin P. M., Guttierrez A. M., Angel S. O., Zaha A., Guarnera E. A. et al. 2005 High polymorphism in genes encoding antigen B from human infections strains of Echinococcus granulosus. Parasitology 131, 805–815.
Kennedy D. A. and Read A. F. 2017 Why does drug resistance readily evolve but vaccine resistance does not? Proc. Biol. Sci. 284, 20162562.
Larrieu E., Mujica G., Gauci C. G., Vizcaychipi K., Seleiman M., Herrero E. et al. 2015 Pilot field trial of the EG95 vaccine against ovine cystic echinococcosis in Rio Negro, Argentina: second study of impact. PLoS Negl. Trop. Dis. 9, e0004134.
Leinikki P., Lehtinen M., Hyoty H., Parkkonen P., Kantanen M. L. and Hakulinen J. 1993 Synthetic peptides as diagnostic tools in virology. Adv. Virus Res. 42, 149–186.
Li Y., Liu X., Zhu Y., Zhou X., Cao C., Hu X. et al. 2013 Bioinformatic prediction of epitopes in the Emy162 antigen of Echinococcus multilocularis. Exp. Ther. Med. 6, 335–340.
Lightowlers M. W. and Heath D. D. 2004 Immunity and vaccine control of Echinnococuss granulosus infection in animal intermediate hosts. Parassitologia 46, 27–31.
Lightowlers M. W., Jensen O. and Fernandez E. 1999 Vaccination trails in Australia and Argentina confirm the effectiveness of the EG95 hydatid vaccine in sheep. Int. J. Parasitol. 29, 531–534.
Lightowlers M. W., Lawrence S. B., Gauci C. G., Young J., Ralston M. J., Maas D. et al. 1996 Vaccination against hydatidosis using a defined recombinant antigen. Parasite Immunol. 18, 457–462.
Lightowlers M. W., Gauci C. G., Chow C., Drew D. R., Gauci S. M., Jenkins D. J. et al. 2003 Molecular and genetic characterization of the host-protective oncospheres antigens of taeniid cestode parasites. Int. J. Parasitol. 33, 1207–1217.
Nono J. K., Pletinckx K., Lutz M. B. and Brehm K. 2012 Excretory/secretory-products of Echinococcus multilocularis Larvae induce apoptosis and tolerogenic properties in dendritic cells in vitro. PLoS Negl Trop Dis. 6, e1516.
Nunnari G., Pinzone M. R., Gruttadauria S., Celesia B. M., Madeddu G., Malaguarnera G. et al. 2012 Hepatic echinococcosis: clinical and therapeutic aspects. World J. Gastroenterol. 18, 1448–1458.
Pan D., Bera A. K., Bandyopadhyay S., Das S. K., Bandyopadhyay S., Bhattacharyya S. et al. 2009 Relative expression of antigen B coding gene of bubaline isolates of Echinococcus granulosus in fertile and sterile cysts. J. Helminthol. 28, 1–4.
Pan D., Bera A. K., Das S. K., Bandyopadhyay S., Manna B. and Bhattacharya D. 2010a Polymorphism and natural selection of antigen B1 of Echinococcus granulosus isolated from different host assemblages in India. Mol. Biol. Rep. 37, 1477–1482.
Pan D., Bera A. K, De S., Bandyopadhyay S., Das S. K., Manna B. et al. 2010b Relative expression of the 14-3-3 gene in different morphotypes of cysts of Echinococcus granulosus isolated from the India buffalo. J. Helminthol. 15, 1–4.
Pellequer J. L., Westhof E. and Van Regenmortel M. H. 1991 Predicting location of continuous epitopes in proteins from their primary structures. Methods Enzymol. 203, 176–201.
Regenmortel M. H. V. V. 2006 Immunoinformatics may lead to a reappraisal of the nature of B cell epitopes and of the feasibility of synthetic peptide vaccines. J. Mol. Recognit. 19, 183–187.
Rosenzvit M. C., Camica F., Kamenentzky L., Muzulin P. M. and Gutierrez A. M 2006 Identification and intra-specific variability analysis of secreted membrane-bound proteins from Echinococcus granulosus. Parasitol Int. 55, 63–67.
Saha S. and Raghava G. P. 2006 Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins 65, 40–48.
Singh H. and Raghava G. P. 2001 Propred: prediction of HLA-DR binding sites. Bioinformatics 7, 1236–1237.
Spotin A., Mahami-Oskouei M., Harandi M. F., Baratchian M., Bordbar A., Ahmadpour E. et al. 2017 Genetic variability of Echinococcus granulosus complex in various geographical populations of Iran inferred by mitochondrial DNA sequences. Acta Trop. 165, 10–16.
Spouge J. L., Guy H. R., Cornette J. L., Margalit H., Cease K., Berzofsky J. A. et al. 1987 Strong conformational propensities enhance T cell antigenicity. J. Immunol. 138, 204–212.
Tamura K., Dudley J., Nei M. and Kumar S. 2007 MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599.
Thompson R. C. A., Lymbery A. J. and Constantine C. C. 1995 Variation in Echinococcus: towards a taxonomic revision of the genus. Adv. Parasitol. 35, 145–175.
Thompson R. C. A. and McManus D. P. 2001 Aetiology: parasites and life-cycles. In WHO/OIE manual on echinococcosis in humans and animals: A public health problem of global concern (ed. J. Eckert, M. A. Gemmel, F. X. Meslin, Z. S. Pawlowski), pp. 1–9. World Organization for Animal Health, Paris.
Torgerson P. R. and Heath D. D. 2003 Transmission dynamics and control options for Echinococcus granulosus. Parasitology 127, 43–58.
WHO (2018) Echinococcosis. Who.int./news-rooms (accessed on 14-01-2019).
WHO-Informal Working Group on Echinococcosis 2003 PAIR: Puncture, aspiration, injection, Re-aspiration. An option for the treatment of cystic echinococcosis. WHO, Geneva.
Woollard D. J., Heath D. D. and Lightowlers M. W. 2000 Assessment of protective immune responses against hydatid disease in sheep by immunization with synthetic peptide antigens. Parasitology 121, 145–153.
Zhang J. 2008 Positive selection, not negative selection, in the pseudogenization of rcsA in Yersinia pestis. Proc. Natl. Acad. Sci. USA 105, E69; author reply E70.
Zhang W., Lij Y. H., Zhang Z., Turson G., Aili H., Wang J. et al. 2003 Echinococcus granulosus from Xinjiang, PR China cDNAs encoding the EG95 vaccine antigen are expressed in different life cycle stages and are conserved in the oncosphere. Am. J. Trop. Med. Hyg. 68, 40–43.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: H. A. Ranganath
Rights and permissions
About this article
Cite this article
Sreevatsava, V., De, S., Bandyopadhyay, S. et al. Variability of the EG95 antigen-coding gene of Echinococcus granulosus in animal and human origin: implications for vaccine development. J Genet 98, 53 (2019). https://doi.org/10.1007/s12041-019-1097-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-019-1097-y