Abstract
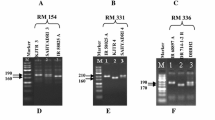
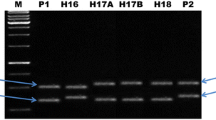
Genomic simple sequence repeat (SSR) markers were used to fingerprint and determine genetic similarity (GS) of the watermelon breeding lines, as well as the purity of their hybrid derivatives. Cluster analysis and Jaccard’s distance coefficients using the unweighted pair group method with arithmetic mean (UPGMA) have classified these lines into three major groups. Notwithstanding, the genetic background of these lines is narrow as revealed by the restricted GS coefficients. Fifty-five sets of SSR markers were employed in this study. Fourteen of these markers were polymorphic between the breeding lines and were used for assessing hybrid purity. Cross-checking assay validated nine SSR markers as informative SSR markers for purity detection of these hybrids. To confirm the accuracy and efficiency of these markers, their derived PCR products were further sequenced, and ClSSR09643, ClSSR18153 and ClSSR01623 were selected as high-efficiency SSR markers. Interestingly, SSR markers ClSSR09643 and ClSSR18153 were broadly applied for purity detection of more than two different hybrids, while SSR marker ClSSR01623 behaved as a specific marker for purity detection in this study. Genetic purity of six commercial watermelon hybrids was definitely evaluated using these SSR markers. Genetic purity of all tested hybrids exceeded 96% while the field purity was above 98%. Genetic purity test was an emergency for identifying off-types and selfed female in a lot of hybrid seeds. Here, we elucidated the potential of nine SSR markers including three with higher breeding selection efficiency. We recommended them to seed company for purity improvement of watermelon commercial hybrid varieties.




Similar content being viewed by others
References
Anderson J. A., Churchill G. A., Autrique J. E. and Sorrell M. E. 1993 Optimizing parental selection for genetic linkage maps. Genome 36, 181–186.
Bo K., Ma Z., Chen J. and Weng Y. 2015 Molecular mapping reveals structural rearrangements and quantitative trait loci underlying traits with local adaptation in semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis Qi et Yuan). Theor. Appl. Genet. 128, 25–39.
Bruford M. W. and Wayne R. K. 1993 Microsatellites and their application to population genetic studies. Curr. Opin. Genet. Dev. 3, 939–943.
Bussell J. D., Waycott M. and Chappill J. A. 2005 Arbitrarily amplified DNA markers as characters for phylogenetic inference. Perspect. Plant Ecol. Evol. Syst. 7, 3–26.
Che K. P., Liang C. Y., Wang Y. G., Jin D. M. and Wang B. 2003 Genetic assessment of watermelon germplasm using the AFLP technique. Hort. Sci. 38, 81–84.
Ding Q. Y. and Zhang X. 2005 Application of molecular marker technique in watermelon and melon. J. Fruit Sci. 03. 271–275.
Garke C., Ytournel F., Bed’hom B., Gut I., Lathrop M., Weigend S. and Simianer H. 2012 Comparison of SNPs and microsatellites for assessing the genetic structure of chicken populations. Anim. Genet. 43, 419–428.
Gort G., Koopman W. J. M., Stein A. and Eeuwijk F. A. 2008 Collision probabilities for AFLP bands, with an application to simple measures of genetic similarity. JABES 13, 177–198.
Guerra-Sanz J. M. 2002 Citrullus simple sequence repeats markers from sequence databases. Mol. Ecol. Notes 2, 223–225.
Guo S. G., Zhang J. G., Sun H. H., Jerome S., William J. L. and Zhang H. Y. 2013 The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. 45. Nat. Genet. 1. https://doi.org/10.1038/ng.2470.
Hamblin M. T., Warburton M. L. and Buckler E. S. 2007 Empirical comparison of simple sequence repeats and single nucleotide polymorphisms in assessment of maize diversity and relatedness. PLoS One 2, e1367.
Hu J. B., Zhou X. Y. and Li J. W. 2010 Development of novel EST-SSR markers for cucumber (Cucumis sativus) and their transferability to related species. Sci. Hort. 125, 534–538.
Jaccard P. 1908 Nouvelles recherché sur la distribution florale. Bull. de la SociétéVaudoise des Sci. Naturelles 44, 223–270 (In French).
Jackson T. F. H. G. and Ravdin J. I. 1996 Differentiation of Entamoeba histolytica and Entamoeba dispar infections. Parasitol. Today 12, 406.
Jarret R. L., Merrick L. C., Holms T., Evans J. and Aradhya M. K. 1996 Simple sequence repeats in watermelon (Citrullus lanatus (Thunb) Matsum. and Nakai). Genome 40, 433–441.
Joobeur T., Gusmini G., Zhang X., Levi A., Xu Y., Wehner T. C. et al 2006 Construction of awatermelon BAC library and identification of SSRs anchored to melonor Arabidopsis genomes. Theor. Appl. Genet. 112, 1553–1562.
Kalia R. J., Rai M. K., Kalia S., Singh R. and Dhawan A. K. 2011 Microsatellite markers: an overview of the recent progress in plants. Euphytica 177, 309–334.
Kwon Y. S. 2013 Use of EST-SSR markers for genetic characterization of commercial watermelon varieties and hybrid seed purity testing. Seed Sci. Technol. 41, 245–256.
Kwon Y. S., Oh Y. H., Yi S. I., Kim H. Y., An J. M., Yang S. G. et al 2010 Informative SSR markers for commercial variety discrimination in watermelon (Citrullus lanatus). Genes and Genomics 32, 115–122.
Lee S. J., Shin J. S., Park K. W. and Hong Y. P. 1996 Detection of genetic diversity using RAPD-PCR and sugar analysis in watermelon (Citrullus lanatus (Thunb.)Mansf. germplasm. Theor. Appl. Genet. 92, 719–725.
Levi A.,Wechter W. P., Davis A. R., Katzir N., Tadmor K., Ling K. et al 2007 Interspecific transferability of watermelon EST-SSR markers in cucurbit species. HortScience 42, 1012.
Levi A., Thies J. A., Wechter W. P., Harrison H. F., Simmons A. M., Reddy U. K. et al 2013 High frequency oligonucleotides: targeting active gene (HFO-TAG) markers revealed wide genetic diversity among Citrullus spp. accessions useful for enhancing disease or pest resistance in watermelon cultivars. Genet. Resour. Crop. Evol. 60, 427–440.
Li C. H., Liu L., Liu X., Zhu L. H., Song R. H., Yang H. J. et al 2015 Seed purity detection and distinctiveness analysis of 5 new watermelon hybrid varieties on SSR markers. CASB 31, 177–185.
Li M. W., Li Z. W., Kong L. C., Wang S. K. and Yao X. F. 2009 Breeding of watermelon hybrid variety ’HuamiGuanlong’. China Cucurbits & Vegetables 22, 16–18.
Li X. H., Wang C. Y. and Chang Z. 2008 Development of molecular markers technique application on genetic breeding of watermelon. Henan Science, http://en.cnki.com.cn/Article_en/CJFDTotal-HNKX200805017.htm.
Liu Q. H., Wang F. R., Zhang J., Yang J., Li R. Z. and Wang Z. W. 2003 Purity detection of Lumianyan 15 hybrid with SSR markers. Shandong Agric. Sci., http://en.cnki.com.cn/Article_en/CJFDTOTAL-AGRI200302002.htm.
Liu G., Liu L., Gong Y., Wang Y., Yu F., Shen H. et al 2007 Seed genetic purity testing of F1 hybrid cabbage (Brassica oleracea var. capitata) with molecular marker analysis. Seed Sci. Technol. 35, 477–486.
Liu J. L., Zhou S. C., Song X. H. and Wang J. S. 2006 Purity test of Melon hybrid with SSR molecular markers. Mol. Plant Breed., http://en.cnki.com.cn/Article_en/CJFDTotal-FZZW2006S2005.htm.
Liu L. W., Hou X. L., Gong Y. Q., Zhang Y. M., Wang K. R. and Zheng J. F. 2004 Application of molecular marker in variety identification and purity testing in vegetable crops. Mol. Plant Breed., http://en.cnki.com.cn/Article_en/CJFDTotal-GSNK201705021.htm.
Lv J., Qi J., Shi Q., Shen D., Zhang S., Shao G. et al 2012 Genetic diversity and population structure of cucumber (Cucumis sativus L. PLoS One 7, e46919.
Ma D. W., Gao S. Z. and Wang Z. J. 1986 A study on peroxidase isozymes in Cucumis melo L. J. Agric. Univ. Hebei, http://agris.fao.org/agris-search/search.do?recordID=CN8601305.
Mei M. and Lu L. 2005 Application of DNA molecular markers technique on quality testing of crop seed. Mol. Plant Breed., http://en.cnki.com.cn/Article_en/CJFDTOTAL-FZZW200501027.htm.
Miao R. Y., Yue Q., Zhi H. Y., Ma H. L. and Liang Y. P. 2010 A new F1 hybrid watermelon – Jinkang Zaoguan. China Cucurbits Vegetable, 10, http://xueshu.baidu.com/s?wd=paperuri.
Mujaju C., Sehic J., Werlemark G., Garkava-Gustavsson L., Fatih M. and Nybom H. 2010 Genetic diversity in watermelon (Citrullus lanatus) landraces from Zimbabwe revealed by RAPD and SSR markers. Hereditas 147, 142–153.
Naresh V., Yamini K. N., Rajendrakumar P. and Kumar V. D. 2009 EST-SSR marker-based assay for the genetic purity assessment of safflower hybrids. Euphytica 170, 347–353.
Ouyang X. X., Xu Y., Zhang H. Y., Kang G. B. and Wang Y. J. 1999 Rapid identification of hybrid purity in watermelon using random amplified polymorphic DNA (RAPD). J. Agric. Biotechnol, http://en.cnki.com.cn/Article_en/CJFDTOTAL-NYSB199901004.htm.
Pallavi H. M., Gowda R., Vishwanath K., Shadakshari Y. G. and Bhanuprakash K. 2011 Identification of SSR markers for hybridity and seed genetic purity testing in sunflower (Helianthus annuus L.). Seed Sci. Technol. 39, 259–264.
Poczai P., Varga I., Laos M., Cseh A., Bell N., Valkonen J. P. T. et al 2013 Advances in plant gene-targeted and functional markers: a review. Plant Methods 9, 6. https://doi.org/10.1186/1746-4811-9-6.
Rana M. K., Singh S. and Bhat K. V. 2006 RAPD, STMS and ISSR markers for genetic diversity and hybrid seed purity testing in cotton. Seed Sci. Technol. 35, 709–721.
Rohlf F. J. 2000 NTSYS-pc: Numerical taxonomy and multivariate analysis system-version 2.10b. Applied Biostatistics Inc., New York.
Simmons M. P., Zhang L. B., Webb C. T. and Müller K. 2007 A penalty of using anonymous dominant markers (AFLPs, ISSRs, and RAPDs) for phylogenetic inference. Mol. Phylogenet. Evol. 42, 528–542.
Sundaram R. M., Naveenkumar B., Biradar S. K., Balachandran S. M., Mishra B., Ilyasahmed M. et al 2008 Identification of informative SSR markers capable of distinguishing hybrid rice parental lines and their utilization in seed purity assessment. Euphytica 163, 215–224.
Varshney R. K., Garner A. and Sorells M. E. 2005 Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 23, 48–63.
Wang Y. W., Samuels T. D. and Wu Y. Q. 2011 Development of 1030 genomic SSR markers in switchgrass. Theor. Appl. Genet. 122, 677–686.
Zhang X., Wang M., Zhang J. and Jianqiang A. Y. 2002 The new watermelon F1 hybrid variety‘Hongguanlong’. Acta Horticulturae Sinica, http://en.cnki.com.cn/Article_en/CJFDTotal-YYXB200201024.htm.
Zhang X. P. and Wang M. 1989 Isozyme analysis of hybrids and their parents on watermelon (Citrullus lanatus (Thanb.) Mansf. J. Fruit Sci., http://en.cnki.com.cn/Article_en/CJFDTOTAL-GSKK198902005.htm.
Zhu H. Y., Song P. Y., Koo D. H., Guo L. Q., Li Y. M., Sun S. R. et al 2016a Genome wide characterization of simple sequence repeats in watermelon genome and their application in comparative mapping and genetic diversity analysis. BMC Genomics 17, 557. https://doi.org/10.1186/s12864-016-2870-4.
Zhu L., Wu L. and Tong G. 1992 Study on the peroxidase isozymes of watermelon hybrids and their parents. J. Anhui Agric. Univ., http://en.cnki.com.cn/Article_en/CJFDTotal-ANHU199204003.htm.
Zhu H., Guo L., Song P., Luan F. and Hu J. 2016b Development of genome-wide SSR markers in melon with their cross-species transferability analysis and utilization in genetic diversity study. Mol. Breed. 36, 153.
Acknowledgements
We are thankful to the Chinese leaders for establishing and promoting Talents projects in Jiangsu Province and Suqian city. This work is supported by the funding from two talent projects ‘2017 Jiangsu province Entrepreneurial innovation talent program’ and ‘2017 Suqian city Entrepreneurial innovation leading talent program’.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Corresponding editor: H. A. Ranganath
Xia Lu and Yawo Mawunyo Nevame Adedze contributed equally to this work. AYMN, SLT and LWH designed the project. LX, AYMN, DZJ, TLH and ZXL conducted the field experiments. LX and AYMN performed the laboratory tests. AYMN and LX analysed the data. AYMN drafted the manuscript. AYMN and LX worked on figures and tables presentation and rearrangement. CGN and GM proof read the manuscript and conducted English editing. CGN performed a critical revision of the article. All authors read and approved the final manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lu, X., Adedze, Y.M.N., Chofong, G.N. et al. Identification of high-efficiency SSR markers for assessing watermelon genetic purity. J Genet 97, 1295–1306 (2018). https://doi.org/10.1007/s12041-018-1027-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-018-1027-4