Abstract
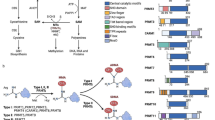
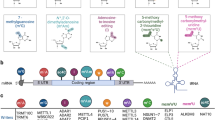
S-adenosyl-L-methionine (AdoMet)-dependent methyltransferases (MTases) are involved in diverse cellular functions. These enzymes show little sequence conservation but have a conserved structural fold. The DNA MTases have characteristic motifs that are involved in AdoMet binding, DNA target recognition and catalysis. Motif III of these MTases have a highly conserved acidic residue, often an aspartate, whose functional significance is not clear. Here, we report a mutational study of the residue in the β family MTase of the Type III restriction-modification enzyme EcoP15I. Replacement of this residue by alanine affects its methylation activity. We propose that this residue contributes to the affinity of the enzyme for AdoMet. Analysis of the structures of DNA, RNA and protein MTases reveal that the acidic residue is conserved in all of them, and interacts with N6 of the adenine moiety of AdoMet. Interestingly, in the SET-domain protein lysine MTases, which have a fold different from other AdoMet-dependent MTases, N6 of the adenine moiety is hydrogen bonded to the main chain carbonyl group of the histidine residue of the highly conserved motif III. Our study reveals the evolutionary conservation of a carbonyl group in DNA, RNA and protein AdoMet-dependent MTases for specific interaction by hydrogen bond with AdoMet, despite the lack of overall sequence conservation.






Similar content being viewed by others
References
Ahmad I, Kulkarni M, Gopinath A and Kayarat S 2018 Single-site DNA cleavage by Type III restriction endonuclease requires a site-bound enzyme and a trans-acting enzyme that are ATPase-activated. Nucl. Acids Res. 46 6229–6237
Ashkenazy H, Erez E, Martz E, Pupko T and Ben-Tal N 2010 ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucl. Acids Res. 38 W529–W533
Atack JM, Murphy TF, Bakaletz, LO, Seib KL and Jennings MP 2018 Closed complete genome sequences of two nontypeableHaemophilusinfluenzae strains containing novel moda alleles from the sputum of patients with chronic obstructive pulmonary disease. Microbiol. Resour. Announc. 7 e00821–e00818
Bheemanaik S, Reddy YV and Rao DN 2006 Structure, function and mechanism of exocyclic DNA methyltransferases. Biochem. J. 399 177–190
Bhujbalrao R and Anand R 2019 Deciphering determinants in ribosomal methyltransferases that confer antimicrobial resistance. J. Am. Chem. Soc. 141 1425–1429
Cheng X, Collins RE, and Zhang X 2005 Structural and sequence motifs of protein (histone) methylation enzymes. Annu. Rev. Biophys. Biomol. Struct. 34 267–294
Cheng X and Roberts RJ 2001 AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucl. Acids Res. 29 3784–3795
Dor Y and Cedar H 2018 Principles of DNA methylation and their implications for biology and medicine. Lancet 392 777–786
Estève PO, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR and Pradhan S 2011 A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat. Struct. Mol. Biol. 18 42
Gong W, O’Gara M, Blumenthal RM, and Cheng X 1997 Structure of pvu II DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucl. Acids Res. 25 2702–2715
Greer EL and Shi Y 2012 Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13 343–357
Gupta YK, Chan SH, Xu SY and Aggarwal AK 2015 Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I. Nat. Commun. 6 7363
Klimašauskas S, Kumar S, Roberts RJ and Cheng X 1994 HhaI methyltransferase flips its target base out of the DNA helix. Cell 76 357–369
Liu, R-J, Long T, Li J, Li H and Wang, E-D 2017 Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6. Nucl. Acids Res. 45 6684–6697
Martin JL and McMillan FM 2002 SAM (dependent) I AM: the S-adenosyl methionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12 783–793
Min J, Feng Q, Li Z, Zhang Y and Xu RM 2003 Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell 112 711–723
Raghavendra NK and Rao DN 2005 Exogenous AdoMet and its analogue sinefungin differentially influence DNA cleavage by R.EcoP15I- usefulness in SAGE. Biochem. Biophys. Res. Commun. 334 803–811
Rao DN, Dryden DTF and Bheemanaik S 2014 Type III restriction-modification enzymes: a historical perspective. Nucl. Acids Res. 42 45–55
Robertson KD 2001 DNA methylation, methyltransferases, and cancer. Oncogene 20 3139–3155
Roth M, Helm-Kruse S, Friedrich T and Jeltsch A 1998 Functional roles of conserved amino acid residues in DNA methyltransferases investigated by site-directed mutagenesis of the EcoRV adenine-N 6-methyltransferase. J. Biol. Chem. 273 17333–17342
Schluckebier G, Kozak M, Bleimling N, Weinhold E and Saenger W 1997 Differential binding of S-adenosylmethionine S-adenosylhomocysteine and Sinefungin to the adenine-specific DNA methyltransferase M. TaqI. J. Mol. Biol. 265 56–67
Thomas CB, Scavetta RD, Gumport RI and Churchill ME 2003 Structures of liganded and unligandedRsrI N6-adenine DNA methyltransferase a distinct orientation for active cofactor binding. J. Biol. Chem. 278 26094–26101
Tran PH, Korszun ZR, Cerritelli S, Springhorn SS and Lacks SA 1998 Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of Streptococcus pneumoniae bound to S-adenosylmethionine. Structure 6 1563–1575
Vasu K and Nagaraja V 2013 Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 77 53–72
Wang M, Zhu Y, Wang C, Fan X, Jiang X, Ebrahimi M, Qiao Z, Niu L, Teng, M and Li. X 2016 Crystal structure of the two-subunit tRNA m1A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae. Sci. Rep. 6 32562
Zhang X and Cheng X 2003 Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure 11 509–520
Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU and Cheng X 2003 Structural basis for the product specificity of histone lysine methyltransferases. Mol. Cell 12 177–185
Acknowledgements
This work was supported by the Science and Engineering Research Board, Department of Science and Technology (DST), India (EMR/2016/000872 to KS). AG thanks DST India for an INSPIRE Scholarship. IA thanks University Grants Commission, India, for a Senior Research Fellowship. OPC thanks the Department of Biotechnology, India, for Research Associate Fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection: Chromatin Biology and Epigenetics.
Rights and permissions
About this article
Cite this article
Gopinath, A., Kulkarni, M., Ahmed, I. et al. The conserved aspartate in motif III of β family AdoMet-dependent DNA methyltransferase is important for methylation. J Biosci 45, 10 (2020). https://doi.org/10.1007/s12038-019-9983-2
Published:
DOI: https://doi.org/10.1007/s12038-019-9983-2