Abstract
Parkinson’s disease (PD) is a chronic and progressive neurodegenerative disorder. While most PD cases are idiopathic, the known genetic causes of PD are useful to understand common disease mechanisms. Recent data suggests that autophagy is regulated by protein acetylation mediated by histone acetyltransferase (HAT) and histone deacetylase (HDAC) activities. The changes in histone acetylation reported to be involved in PD pathogenesis have prompted this investigation of protein acetylation and HAT and HDAC activities in both idiopathic PD and G2019S leucine-rich repeat kinase 2 (LRRK2) cell cultures. Fibroblasts from PD patients (with or without the G2019S LRRK2 mutation) and control subjects were used to assess the different phenotypes between idiopathic and genetic PD. G2019S LRRK2 mutation displays increased mitophagy due to the activation of class III HDACs whereas idiopathic PD exhibits downregulation of clearance of defective mitochondria. This reduction of mitophagy is accompanied by more reactive oxygen species (ROS). In parallel, the acetylation protein levels of idiopathic and genetic individuals are different due to an upregulation in class I and II HDACs. Despite this upregulation, the total HDAC activity is decreased in idiopathic PD and the total HAT activity does not significantly vary. Mitophagy upregulation is beneficial for reducing the ROS-induced harm in genetic PD. The defective mitophagy in idiopathic PD is inherent to the decrease in class III HDACs. Thus, there is an imbalance between total HATs and HDACs activities in idiopathic PD, which increases cell death. The inhibition of HATs in idiopathic PD cells displays a cytoprotective effect.






Similar content being viewed by others
Abbreviations
- AA:
-
anacardic acid
- Ac-K:
-
acetylated lysine
- ATG:
-
autophagy-related
- BAF. A1:
-
bafilomycin A1
- CBP:
-
CREB-binding protein
- CCCP:
-
carbonyl cyanide 3-chlorophenylhydrazone
- COX IV:
-
cytochrome c oxidase subunit 4
- CsA:
-
cyclosporin A
- DiOC6(3):
-
3,3′-dihexyloxacarbocyanine iodide
- DRP1:
-
dynamin related protein 1
- EBSS:
-
Earle’s balanced salt solution
- ERK1/2:
-
extracellular signal-regulated kinase 1/2
- GAPDH:
-
glyceraldehyde-3-phosphate dehydrogenase
- GNAT:
-
GCN5-related N-acetyltransferase
- GFP:
-
green fluorescent protein
- H3:
-
histone 3
- H3K14:
-
histone 3 lysine 14
- H4:
-
histone 4
- H4K5K8K12:
-
histone 4 lysine 5 lysine 8 lysine 12
- H4K16:
-
histone 4 lysine 16
- HAT:
-
histone acetyltransferase
- HDAC:
-
histone deacetylase
- HFs:
-
human fibroblasts
- hMOF:
-
human male absent of first
- IPD:
-
idiopathic Parkinson’s disease
- JNK1:
-
Jun-N-terminal kinase 1
- LC3:
-
light-chain microtubule-associated protein
- LONP1:
-
lon peptidase 1
- LRRK2:
-
leucine-rich repeat kinase 2
- MAPK:
-
mitogen-activated protein kinase
- MMP:
-
mitochondrial membrane potential
- MPP+ :
-
1-methyl-4-phenylpyridinium iodide
- MPTP:
-
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine hydrochloride
- MTG:
-
MitoTracker green
- mTOR:
-
mammalian target of rapamycin
- NAD+ :
-
nicotinamide adenine dinucleotide
- NAM:
-
nicotinamide
- PCAF:
-
p300/CREB-binding protein-associated factor
- PD:
-
Parkinson’s disease
- PI:
-
propidium iodide
- PINK1:
-
PTEN-induced putative kinase 1
- ROS:
-
reactive oxygen species
- RT:
-
room temperature
- SIRT:
-
sirtuin
- TIP60:
-
tat-interactive protein 60
- TMRM:
-
tetramethylrhodamine methyl ester perchlorate
- TSA:
-
trichostatin A
- WB:
-
western blotting
- WT:
-
wild-type
References
Lill CM (2016) Genetics of Parkinson’s disease. Mol Cell Probes 30(6):386–396
Niso-Santano M, González-Polo RA, Bravo-San Pedro JM, Gómez-Sánchez R, Lastres-Becker I, Ortiz-Ortiz MA, Soler G, Morán JM et al (2010) Activation of apoptosis signal-regulating kinase 1 is a key factor in paraquat-induced cell death: modulation by the Nrf2/Trx axis. Free Radic Biol Med 48(10):1370–1381
Gonzalez-Polo RA et al (2004) Paraquat-induced apoptotic cell death in cerebellar granule cells. Brain Res 1011(2):170–176
Song C, Kanthasamy A, Anantharam V, Sun F, Kanthasamy AG (2010) Environmental neurotoxic pesticide increases histone acetylation to promote apoptosis in dopaminergic neuronal cells: relevance to epigenetic mechanisms of neurodegeneration. Mol Pharmacol 77(4):621–632
Bernstein, A.I., et al., 6-OHDA generated ROS induces DNA damage and p53- and PUMA-dependent cell death. Mol Neurodegener, 2011. 6(1): p. 2.
Jin SM, Youle RJ (2013) The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy 9(11):1750–1757
Gomez-Sanchez R et al (2016) PINK1 deficiency enhances autophagy and mitophagy induction. Mol Cell Oncol 3(2):e1046579
Gonzalez-Polo RA, Niso-Santano M, Ortiz-Ortiz MA, Gomez-Martin A, Moran JM, Garcia-Rubio L, Francisco-Morcillo J, Zaragoza C et al (2007) Inhibition of paraquat-induced autophagy accelerates the apoptotic cell death in neuroblastoma SH-SY5Y cells. Toxicol Sci 97(2):448–458
Gonzalez-Polo RA et al (2015) Is the modulation of autophagy the future in the treatment of neurodegenerative diseases? Curr Top Med Chem 15(21):2152–2174
Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J, Ma C, Sun Y et al (2012) Function and molecular mechanism of acetylation in autophagy regulation. Science 336(6080):474–477
Sterner DE, Berger SL (2000) Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 64(2):435–459
Costantini S, Sharma A, Raucci R, Costantini M, Autiero I, Colonna G (2013) Genealogy of an ancient protein family: the Sirtuins, a family of disordered members. BMC Evol Biol 13:60
Delcuve GP, Khan DH, Davie JR (2012) Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics 4(1):5
Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, Zhang SM, Lian G et al (2012) GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 336(6080):477–481
Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW et al (2008) A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A 105(9):3374–3379
Lee IH, Finkel T (2009) Regulation of autophagy by the p300 acetyltransferase. J Biol Chem 284(10):6322–6328
Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM et al (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12(1):1–222
Marino G et al (2014) Regulation of autophagy by cytosolic acetyl-coenzyme a. Mol Cell 53(5):710–725
Jowaed A, Schmitt I, Kaut O, Wullner U (2010) Methylation regulates alpha-synuclein expression and is decreased in Parkinson's disease patients' brains. J Neurosci 30(18):6355–6359
Song C, Kanthasamy A, Jin H, Anantharam V, Kanthasamy AG (2011) Paraquat induces epigenetic changes by promoting histone acetylation in cell culture models of dopaminergic degeneration. Neurotoxicology 32(5):586–595
Goers J, Manning-Bog AB, McCormack AL, Millett IS, Doniach S, di Monte DA, Uversky VN, Fink AL (2003) Nuclear localization of alpha-synuclein and its interaction with histones. Biochemistry 42(28):8465–8471
Kontopoulos E, Parvin JD, Feany MB (2006) Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet 15(20):3012–3023
Monti B, Gatta V, Piretti F, Raffaelli SS, Virgili M, Contestabile A (2010) Valproic acid is neuroprotective in the rotenone rat model of Parkinson’s disease: involvement of alpha-synuclein. Neurotox Res 17(2):130–141
Yakhine-Diop SM et al (2014) G2019S LRRK2 mutant fibroblasts from Parkinson's disease patients show increased sensitivity to neurotoxin 1-methyl-4-phenylpyridinium dependent of autophagy. Toxicology 324:1–9
Papkovskaia TD, Chau KY, Inesta-Vaquera F, Papkovsky DB, Healy DG, Nishio K, Staddon J, Duchen MR et al (2012) G2019S leucine-rich repeat kinase 2 causes uncoupling protein-mediated mitochondrial depolarization. Hum Mol Genet 21(19):4201–4213
Riss, T.L., et al., Cell Viability Assays, in Assay Guidance Manual, G.S. Sittampalam, et al., Editors. 2004: Bethesda (MD).
Rodriguez-Arribas M et al (2017) Turnover of lipidated LC3 and autophagic cargoes in mammalian cells. Methods Enzymol 587:55–70
Mauro-Lizcano M, Esteban-Martínez L, Seco E, Serrano-Puebla A, Garcia-Ledo L, Figueiredo-Pereira C, Vieira HLA, Boya P (2015) New method to assess mitophagy flux by flow cytometry. Autophagy 11(5):833–843
Robinson KM, Janes MS, Pehar M, Monette JS, Ross MF, Hagen TM, Murphy MP, Beckman JS (2006) Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc Natl Acad Sci U S A 103(41):15038–15043
Pietrocola F, Mariño G, Lissa D, Vacchelli E, Malik SA, Niso-Santano M, Zamzami N, Galluzzi L et al (2012) Pro-autophagic polyphenols reduce the acetylation of cytoplasmic proteins. Cell Cycle 11(20):3851–3860
Yakhine-Diop SM et al (2017) Fluorescent FYVE chimeras to quantify PtdIns3P synthesis during autophagy. Methods Enzymol 587:257–269
Rodriguez-Arribas M et al (2016) IFDOTMETER: a new software application for automated immunofluorescence analysis. J Lab Autom 21(2):246–259
Bolte S, Cordelieres FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224(Pt 3):213–232
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45–e445
Bravo-San Pedro JM, Niso-Santano M, Gómez-Sánchez R, Pizarro-Estrella E, Aiastui-Pujana A, Gorostidi A, Climent V, López de Maturana R et al (2013) The LRRK2 G2019S mutant exacerbates basal autophagy through activation of the MEK/ERK pathway. Cell Mol Life Sci 70(1):121–136
Gomez-Sanchez R et al (2014) Mitochondrial impairment increases FL-PINK1 levels by calcium-dependent gene expression. Neurobiol Dis 62:426–440
Wei X, Qi Y, Zhang X, Qiu Q, Gu X, Tao C, Huang D, Zhang Y (2014) Cadmium induces mitophagy through ROS-mediated PINK1/Parkin pathway. Toxicol Mech Methods 24(7):504–511
Thomas KJ, Jacobson MR (2012) Defects in mitochondrial fission protein dynamin-related protein 1 are linked to apoptotic resistance and autophagy in a lung cancer model. PLoS One 7(9):e45319
Min SW et al (2013) Sirtuins in neurodegenerative diseases: an update on potential mechanisms. Front Aging Neurosci 5:53
Newman JC, He W, Verdin E (2012) Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J Biol Chem 287(51):42436–42443
Su YC, Guo X, Qi X (2015) Threonine 56 phosphorylation of Bcl-2 is required for LRRK2 G2019S-induced mitochondrial depolarization and autophagy. Biochim Biophys Acta 1852(1):12–21
Jin H., et al. -Synuclein negatively regulates protein kinase C expression to suppress apoptosis in dopaminergic neurons by reducing p300 histone acetyltransferase activity. J Neurosci, 2011. 31(6): p. 2035–2051.
Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y (1999) Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem 274(3):1189–1192
de Ruijter AJ et al (2003) Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 370(Pt 3):737–749
Kao HY, Lee CH, Komarov A, Han CC, Evans RM (2002) Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J Biol Chem 277(1):187–193
Green KN, Steffan JS, Martinez-Coria H, Sun X, Schreiber SS, Thompson LM, LaFerla FM (2008) Nicotinamide restores cognition in Alzheimer’s disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J Neurosci 28(45):11500–11510
Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK (2003) Small molecule modulators of histone acetyltransferase p300. J Biol Chem 278(21):19134–19140
Shi S, Lin J, Cai Y, Yu J, Hong H, Ji K, Downey JS, Lu X et al (2014) Dimeric structure of p300/CBP associated factor. BMC Struct Biol 14:2
Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, Liu B, Chang C et al (2015) Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell 57(3):456–466
Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, Bordone L (2009) JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS One 4(12):e8414
Li Y, Xu W, McBurney MW, Longo VD (2008) SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab 8(1):38–48
Zhao Y, Luo P, Guo Q, Li S, Zhang L, Zhao M, Xu H, Yang Y et al (2012) Interactions between SIRT1 and MAPK/ERK regulate neuronal apoptosis induced by traumatic brain injury in vitro and in vivo. Exp Neurol 237(2):489–498
Dai SH, Chen T, Wang YH, Zhu J, Luo P, Rao W, Yang YF, Fei Z et al (2014) Sirt3 protects cortical neurons against oxidative stress via regulating mitochondrial Ca2+ and mitochondrial biogenesis. Int J Mol Sci 15(8):14591–14609
Liu L, Peritore C, Ginsberg J, Shih J, Arun S, Donmez G (2015) Protective role of SIRT5 against motor deficit and dopaminergic degeneration in MPTP-induced mice model of Parkinson’s disease. Behav Brain Res 281:215–221
Zhang JY, Deng YN, Zhang M, Su H, Qu QM (2016) SIRT3 acts as a neuroprotective agent in rotenone-induced Parkinson cell model. Neurochem Res 41(7):1761–1773
Yakhine-Diop SMS et al Acetylome in human fibroblasts from Parkinson’s disease patients. Front Cell Neurosci 2018, 12:97
Rouaux C, Jokic N, Mbebi C, Boutillier S, Loeffler JP, Boutillier AL (2003) Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J 22(24):6537–6549
Choong CJ, Sasaki T, Hayakawa H, Yasuda T, Baba K, Hirata Y, Uesato S, Mochizuki H (2016) A novel histone deacetylase 1 and 2 isoform-specific inhibitor alleviates experimental Parkinson’s disease. Neurobiol Aging 37:103–116
Salama AF et al (2015) Epigenetic study of Parkinson’s disease in experimental animal model. International Journal of Clinical and Experimental Neurology 3(1):11–20
More SV, Choi DK (2016) Emerging preclinical pharmacological targets for Parkinson’s disease. Oncotarget 7(20):29835–29863
Schapira AH et al (1990) Mitochondrial complex I deficiency in Parkinson’s disease. J Neurochem 54(3):823–827
Acknowledgments
We are grateful to the patients and donors without whom this work would not have been possible. The authors thank M. P. Delgado-Luceño and Dr. J.A. Tapia-Garcia. The authors also thank FUNDESALUD for helpful assistance.
Funding
SMS.Y-D was supported by Isabel Gemio Foundation. M. N-S was funded by “Ramon y Cajal Program (RYC-2016-20883) Spain. M. R-A. and E. U-C were supported by a FPU predoctoral fellowship (FPU13/01237 and FPU16/00684, respectively) from Ministerio de Educación, Cultura y Deporte, Spain. R. G-S. was supported by a Marie Sklodowska-Curie Individual Fellowship (IF-EF) (655027) from the European Commission. JM. B-S. P. was funded by La Ligue Contre le Cancer. JM. F. received research support from the Instituto de Salud Carlos III, CIBERNED (CB06/05/004) and Instituto de Salud Carlos III, FIS, (PI15/00034). RA. G-P. was supported by a “Contrato destinado a la retención y atracción del talento investigador, TA13009“ from Junta de Extremadura, and received a research support from the Instituto de Salud Carlos III, FIS, (PI14/00170). JM.C. was funded by a Parkinson’s UK project grant (G-1406). This work was also supported by “Fondo Europeo de Desarrollo Regional” (FEDER) from the European Union.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
ESM 1
(DOCX 12 kb)
ESM 2
(DOCX 13 kb)
Figure S1
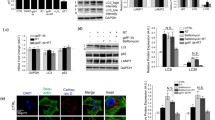
Determination of G2019S LRRK2 mutation and expression levels of SIRT2 and SIRT6. A/ Restriction enzyme of LRRK2 exon 41. Bfm I hydrolyses the exon 41 harboring the G2019S mutation into 2 bands (300 and 200 base pairs (bp)) confirming that the mutation is heterozygous. SIRT2 expression levels. B/ mRNA expression of SIRT2 by qPCR. Data are the normalized mean ± SD of three independent experiments. C-E/ Detection of two isoforms of SIRT2 by immunoblotting, SIRT2 isoform I (43 kDa) (C, D) and SIRT2 isoform II (39 kDa) (C, E). The densitometry of each isoform is normalized to GAPDH. The results correspond to the relative mean ± SD of three independent experiments, *p < 0.05 in comparison to Co, (Student’s t-test). F-H/ SIRT6 expression levels. F/ mRNA expression of SIRT6 by qPCR. Data are the normalized mean ± SD of three independent experiments, **p < 0.01 versus Co, (Student’s t-test). G, H/ Assessment of SIRT6 expression and its quantification to referenced GAPDH. Data are the normalized means ± SD of three independent experiments, **p < 0.01 up to Co, (Student’s t-test). (PNG 319 kb)
Figure S2
Ac-K proteins and mRNA expression levels of the HAT family. A, B/ Immunofluorescence intensity of labeled nuclear Ac-K proteins (red) in HFs, the nuclei were stained with Hoechst 33,342 (blue). Original magnification: 40X, scale bar corresponds to 10 μm. B/ Represents the quantification of the fluorescence intensity (n = 60 cells/condition). Data are the mean ± SD of two independent experiments, **p < 0.01, ***p˂0.001 in comparison to Co, (Student’s t-test). p300 (C) and PCAF (D) are members of the p300/CBP and GNAT families, respectively. Members of the MYST family are hMOF (E) and TIP60 (F). Their mRNA expression levels were assessed by qPCR, and the results represent the relative mean ± SD of at least three independent experiments, *p˂0.05, **p˂0.01, ***p˂0.001 versus Co, (Student’s t-test). G/ Immunofluorescence intensity of labeled cytoplasmic Ac-K proteins (red) in HFs treated with TSA (1 μM) during 4 h. Scale bar corresponds to 10 μm. (PNG 340 kb)
Figure S3
mRNA expression levels of class I and II HDACs. A-C/ Class I HDACs includes HDAC1 (A), HDAC2 (B) and HDAC3 (C). D, E/ Class II HDACs includes HDAC4 (D) and HDAC6 (E). Their mRNA expression levels were assessed by qPCR, and the results correspond to the relative mean ± SD of three independent experiments, *p˂0.05, **p˂0.01, ***p˂0.001 versus Co, (Student’s t-test). (PNG 104 kb)
Figure S4
Cell viability with HDAC inhibitors. A, B/ HFs were treated with TSA (0–100 μM) or NAM (0–100 mM) for 24 h. Cell viability was assessed by the colorimetric test MTT. Data correspond to the normalized mean percentage of untreated cells, *p˂0.05, **p˂0.01, ***p˂0.001, (Student’s t-test). C, D/ Cells were treated with TSA (1 μM) or NAM (1 mM) for 4 h. Histone 3 acetylated on lysine 14 (Ac-H3K14) was detected by immunoblotting and was normalized to total Histone 3 (H3). E/ Cells were treated overnight with EX-527 (1 μM). Cells were stained with propidium iodide (PI), and the percentage of PI+ cells was evaluated by flow cytometry, (n = 10.000 events). Data are the mean percentage ± SD of three independent experiments, *p˂0.05, **p˂0.01, (Student’s t-test). (PNG 236 kb)
Rights and permissions
About this article
Cite this article
Yakhine-Diop, S.M.S., Niso-Santano, M., Rodríguez-Arribas, M. et al. Impaired Mitophagy and Protein Acetylation Levels in Fibroblasts from Parkinson’s Disease Patients. Mol Neurobiol 56, 2466–2481 (2019). https://doi.org/10.1007/s12035-018-1206-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-018-1206-6