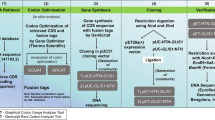
Abstract
The stem cell-specific SOX2 transcription factor is critical for early embryonic development and the maintenance of embryonic and neural stem cell identity. It is also crucial for the generation of induced pluripotent and neural stem cells, thus providing immense prospect in patient-specific therapies. Here, we report soluble expression and purification of human SOX2 protein under native conditions from a bacterial system. To generate this macromolecule, we codon-optimized the protein-coding sequence and fused it to a nuclear localization signal, a protein transduction domain, and a His-tag. This was then cloned into a protein expression vector and was expressed in Escherichia coli. Subsequently, we have screened and identified the optimal expression conditions to obtain recombinant fusion protein in a soluble form and studied its expression concerning the position of fusion tags at either terminal. Furthermore, we purified two versions of recombinant SOX2 fusion proteins to homogeneity under native conditions and demonstrated that they maintained their secondary structure. This molecular tool can substitute genetic and viral forms of SOX2 to facilitate the derivation of integration-free induced pluripotent and neural stem cells. Furthermore, it can be used in elucidating its role in stem cells, various cellular processes and diseases, and for structural and biochemical studies.





Similar content being viewed by others
References
Takahashi, K., & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126, 663–676.
Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science, 318, 1917–1920.
Rowe, R. G., & Daley, G. Q. (2019). Induced pluripotent stem cells in disease modelling and drug discovery. Nature Reviews Genetics, 20, 377–388.
Haridhasapavalan, K. K., Borgohain, M. P., Dey, C., Saha, B., Narayan, G., Kumar, S., & Thummer, R. P. (2019). An insight into non-integrative gene delivery approaches to generate transgene-free induced pluripotent stem cells. Gene, 686, 146–159.
Borgohain, M. P., Haridhasapavalan, K. K., Dey, C., Adhikari, P., & Thummer, R. P. (2019). An insight into DNA-free reprogramming approaches to generate integration-free induced pluripotent stem cells for prospective biomedical applications. Stem Cell Reviews and Reports, 15, 286–313.
Stadtfeld, M., & Hochedlinger, K. (2010). Induced pluripotency: History, mechanisms, and applications. Genes and Development, 24, 2239–2263.
Sommer, C. A., & Mostoslavsky, G. (2013). The evolving field of induced pluripotency: Recent progress and future challenges. Journal of Cellular Physiology, 228, 267–275.
O’Malley, J., Woltjen, K., & Kaji, K. (2009). New strategies to generate induced pluripotent stem cells. Current Opinion in Biotechnology, 20, 516–521.
Dey, C., Narayan, G., Krishna Kumar, H., Borgohain, M. P., Lenka, N., & Thummer, R. P. (2017). Cell-penetrating peptides as a tool to deliver biologically active recombinant proteins to generate transgene-free induced pluripotent stem cells. Studies on Stem Cell Research and Therapy, 3, 6–15.
Seo, B. J., Hong, Y. J., & Do, J. T. (2017). Cellular reprogramming using protein and cell-penetrating peptides. International Journal of Molecular Medicine, 18, 552.
Kaur, J., Kumar, A., & Kaur, J. (2018). Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. Internaional Journal of Biology and Macromolecules, 106, 803–822.
Borgohain, M. P., Narayan, G., Kumar, H. K., Dey, C., & Thummer, R. P. (2018). Maximizing expression and yield of human recombinant proteins from bacterial cell factories for biomedical applications. In P. Kumar, J. K. Patra, & P. Chandra (Eds.), Advances in microbial biotechnology (pp. 447–486). Burlington: Apple Academic Press.
Cho, H.-J., Lee, C.-S., Kwon, Y.-W., Paek, J. S., Lee, S.-H., Hur, J., et al. (2010). Induction of pluripotent stem cells from adult somatic cells by protein-based reprogramming without genetic manipulation. Blood, 116, 386–395.
Zhou, H., Wu, S., Joo, J. Y., Zhu, S., Han, D. W., Lin, T., et al. (2009). Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell, 4, 381–384.
Zhang, H., Ma, Y., Gu, J., Liao, B., Li, J., Wong, J., & Jin, Y. (2012). Reprogramming of somatic cells via TAT-mediated protein transduction of recombinant factors. Biomaterials, 33, 5047–5055.
Nemes, C., Varga, E., Polgar, Z., Klincumhom, N., Pirity, M. K., & Dinnyes, A. (2014). Generation of mouse induced pluripotent stem cells by protein transduction. Tissue Engineering Part C: Methods, 20, 383–392.
Lee, J., Sayed, N., Hunter, A., Au, K. F., Wong, W. H., Mocarski, E. S., et al. (2012). Activation of innate immunity is required for efficient nuclear reprogramming. Cell, 151, 547–558.
Khan, M., Narayanan, K., Lu, H., Choo, Y., Du, C., Wiradharma, N., et al. (2013). Delivery of reprogramming factors into fibroblasts for generation of non-genetic induced pluripotent stem cells using a cationic bolaamphiphile as a non-viral vector. Biomaterials, 34, 5336–5343.
Masui, S., Nakatake, Y., Toyooka, Y., Shimosato, D., Yagi, R., Takahashi, K., et al. (2007). Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nature Cell Biology, 9, 625–635.
Sarkar, A., & Hochedlinger, K. (2013). The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell, 12, 15–30.
Zhang, S., & Cui, W. (2014). Sox2, a key factor in the regulation of pluripotency and neural differentiation. World Journal of Stem Cells, 6, 305–311.
Avilion, A. A., Nicolis, S. K., Pevny, L. H., Perez, L., Vivian, N., & Lovell-Badge, R. (2003). Multipotent cell lineages in early mouse development depend on SOX2 function. Genes and Development, 17, 126–140.
Pevny, L. H., & Nicolis, S. K. (2010). Sox2 roles in neural stem cells. International Journal of Biochemistry and Cell Biology, 42, 421–424.
Adachi, K., Suemori, H., Yasuda, S., Nakatsuji, N., & Kawase, E. (2010). Role of SOX2 in maintaining pluripotency of human embryonic stem cells. Genes to Cells, 15, 455–470.
Haridhasapavalan, K. K., Raina, K., Dey, C., Adhikari, P., & Thummer, R. P. (2020). An insight into reprogramming barriers to iPSC generation. Stem Cells Reviews and Reports, 16, 56–81.
Chuang, H.-M., Huang, M.-H., Chen, Y.-S., & Harn, H.-J. (2020). SOX2 for stem cell therapy and medical use: Pros or cons? Cell Transplantation, 29, 0963689720907565.
Wuebben, E. L., & Rizzino, A. (2017). The dark side of SOX2: cancer-a comprehensive overview. Oncotarget, 8, 44917–44943.
Zhang, S., Xiong, X., & Sun, Y. (2020). Functional characterization of SOX2 as an anticancer target. Signal Transduction and Targeted Therapy, 5, 1–17.
Haridhasapavalan, K. K., Sundaravadivelu, P. K., & Thummer, R. P. (2020). Codon optimization, cloning, expression, purification, and secondary structure determination of human ETS2 transcription factor. Molecular Biotechnology, 62, 485–494.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Micsonai, A., Wien, F., Bulyáki, É., Kun, J., Moussong, É., Lee, Y.-H., et al. (2018). BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Research, 46, W315–W322.
Vasina, J. A., & Baneyx, F. (1997). Expression of aggregation-prone recombinant proteins at low temperatures: A comparative study of the Escherichia coli cspAandtacPromoter Systems. Protein Expression and Purification, 9, 211–218.
Sørensen, H. P., & Mortensen, K. K. (2005). Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microbial Cell Factories, 4, 1.
San-Miguel, T., Pérez-Bermúdez, P., & Gavidia, I. (2013). Production of soluble eukaryotic recombinant proteins in E. coli is favoured in early log-phase cultures induced at low temperature. Springerplus, 2, 89.
Ryan, B. J., & Henehan, G. T. (2013). Overview of approaches to preventing and avoiding proteolysis during expression and purification of proteins. Current Protocols in Protein Science, 71, 5–25.
Huang, C.-J., Lin, H., & Yang, X. (2012). Industrial production of recombinant therapeutics in Escherichia coli and its recent advancements. Journal of Industrial Microbiology and Biotechnology, 39, 383–399.
Bosnali, M., & Edenhofer, F. (2008). Generation of transducible versions of transcription factors Oct4 and Sox2. Biological Chemistry, 389, 851–861.
Pan, C., Lu, B., Chen, H., & Bishop, C. E. (2010). Reprogramming human fibroblasts using HIV-1 TAT recombinant proteins OCT4, SOX2, KLF4 and c-MYC. Molecular Biology Reports, 37, 2117–2124.
Maertens, B., Spriestersbach, A., von Groll, U., Roth, U., Kubicek, J., Gerrits, M., et al. (2010). Gene optimization mechanisms: A multi-gene study reveals a high success rate of full-length human proteins expressed in Escherichia coli. Protein Science, 19, 1312–1326.
Bornhorst, J. A., & Falke, J. J. (2000). Purification of proteins using polyhistidine affinity tags. In J. Thorner, S. D. Emr, & J. N. Abelson (Eds.), Methods in enzymology (Vol. 326, pp. 245–254). Amsterdam: Elsevier.
Greenfield, N. J. (2006). Using circular dichroism spectra to estimate protein secondary structure. Nature Protocol, 1, 2876–2890.
Kelly, S. M., Jess, T. J., & Price, N. C. (2005). How to study proteins by circular dichroism. Biochimica et Biophysica Acta (BBA): Proteins and Proteomics, 1751, 119–139.
Thier, M., Münst, B., Mielke, S., & Edenhofer, F. (2012). Cellular reprogramming employing recombinant sox2 protein. Stem Cells International, 2012, 549846.
Burgess-Brown, N. A., Sharma, S., Sobott, F., Loenarz, C., Oppermann, U., & Gileadi, O. (2008). Codon optimization can improve expression of human genes in Escherichia coli: A multi-gene study. Protein Expression and Purification, 59, 94–102.
Narayan, G., Sundaravadivelu, P. K., Agrawal, A., Gogoi, R., Nagotu, S., & Thummer, R. P. (2020). Soluble expression, purification, and secondary structure determination of human PDX1 transcription factor. Protein Expression and Purification, 180, 105807.
Braun, P., Hu, Y., Shen, B., Halleck, A., Koundinya, M., Harlow, E., & LaBaer, J. (2002). Proteome-scale purification of human proteins from bacteria. Proceedings of the National Academy of Sciences of the United States of America, 99, 2654–2659.
Ghasemi, Y., Ghoshoon, M. B., Taheri, M., Negahdaripour, M., & Nouri, F. (2020). Cloning, expression and purification of human PDGF-BB gene in Escherichia coli: New approach in PDGF-BB protein production. Gene Reports, 19, 100653.
Galluccio, M., Amelio, L., Scalise, M., Pochini, L., Boles, E., & Indiveri, C. (2012). Over-expression in E. coli and purification of the human OCTN2 transport protein. Molecular Biotechnology, 50, 1–7.
Bhat, E. A., Sajjad, N., Sabir, J. S. M., Kamli, M. R., Hakeem, K. R., Rather, I. A., & Bahieldin, A. (2020). Molecular cloning, expression, overproduction and characterization of human TRAIP Leucine zipper protein. Saudi Journal of Biological Sciences, 27, 1562–1565.
Zamani, M., Berenjian, A., Hemmati, S., Nezafat, N., Ghoshoon, M. B., Dabbagh, F., et al. (2015). Cloning, expression, and purification of a synthetic human growth hormone in Escherichia coli using response surface methodology. Molecular Biotechnology, 57, 241–250.
Baneyx, F., & Mujacic, M. (2004). Recombinant protein folding and misfolding in Escherichia coli. Nature Biotechnology, 22, 1399–1408.
Rosano, G. L., & Ceccarelli, E. A. (2014). Recombinant protein expression in Escherichia coli: Advances and challenges. Frontiers in Microbiology, 5, 172.
Vincentelli, R., & Romier, C. (2013). Expression in Escherichia coli: becoming faster and more complex. Current Opinion in Biotechnology, 23, 326–334.
Hu, P. F., Guan, W. J., Li, X. C., & Ma, Y. H. (2012). Construction of recombinant proteins for reprogramming of endangered Luxi cattle fibroblast cells. Molecular Biology Reports, 39, 7175–7182.
Mirakhori, F., Zeynali, B., Rassouli, H., Salekdeh, G. H., & Baharvand, H. (2015). Direct conversion of human fibroblasts into dopaminergic neural progenitor-like cells using TAT-mediated protein transduction of recombinant factors. Biochemical and Biophysical Research Communications, 459, 655–661.
Pouya, A., Rassouli, H., Rezaei-Larijani, M., Salekdeh, G. H., & Baharvand, H. (2020). SOX2 protein transduction directly converts human fibroblasts into oligodendrocyte-like cells. Biochemical and Biophysical Research Communications, 525, 1–7.
Chen, F., Zhang, G., Yu, L., Feng, Y., Li, X., Zhang, Z., et al. (2016). High-efficiency generation of induced pluripotent mesenchymal stem cells from human dermal fibroblasts using recombinant proteins. Stem Cell Research and Therapy, 7, 99.
Tsumoto, K., Ejima, D., Kumagai, I., & Arakawa, T. (2003). Practical considerations in refolding proteins from inclusion bodies. Protein Expression and Purification, 28, 1–8.
Burgess, R. R. (2009). Refolding solubilized inclusion body proteins. In R. R. Burges & M. P. Deutscher (Eds.), Methods in enzymology (Vol. 463, pp. 259–282). Amsterdam: Elsevier.
Ryu, J., Park, H. H., Park, J. H., Lee, H. J., Rhee, W. J., & Park, T. H. (2016). Soluble expression and stability enhancement of transcription factors using 30Kc19 cell-penetrating protein. Applied Microbiology and Biotechnology, 100, 3523–3532.
Araki, Y., Hamafuji, T., Noguchi, C., & Shimizu, N. (2012). Efficient recombinant production in mammalian cells using a novel IR/MAR gene amplification method. PLoS ONE, 7, e41787.
Münst, B., Thier, M. C., Winnemöller, D., Helfen, M., Thummer, R. P., & Edenhofer, F. (2016). Nanog induces suppression of senescence through downregulation of p27KIP1 expression. Journal of Cell Science, 129, 912–920.
Peitz, M., Münst, B., Thummer, R. P., Helfen, M., & Edenhofer, F. (2014). Cell-permeant recombinant Nanog protein promotes pluripotency by inhibiting endodermal specification. Stem Cell Research, 12, 680–689.
Thier, M., Münst, B., & Edenhofer, F. (2010). Exploring refined conditions for reprograming cells by recombinant Oct4 protein. International Journal of Developmental Biology, 54, 1713.
Ring, K. L., Tong, L. M., Balestra, M. E., Javier, R., Andrews-Zwilling, Y., Li, G., et al. (2012). Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell, 11, 100–109.
Acknowledgments
We thank all the members of the Laboratory for Stem Cell Engineering and Regenerative Medicine (SCERM) for their critical reading and excellent support. The authors gratefully acknowledge the support of DBT Program Support (Prof. S.S. Ghosh), Department of Biosciences and Bioengineering, IIT Guwahati, for their assistance in Circular Dichroism experiments. This work was supported by North Eastern Region—Biotechnology Programme Management Cell (NERBPMC), Department of Biotechnology, Government of India (BT/PR16655/NER/95/132/2015), and also by IIT Guwahati Institutional Top-Up on Start-Up Grant.
Author information
Authors and Affiliations
Contributions
MT and CD were responsible for conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, and final approval of the manuscript; SB was responsible for collection and/or assembly of data, data analysis and interpretation, and final editing and approval of the manuscript; SS was responsible for conception and design, data analysis and interpretation, final approval of manuscript and financial support; and RPT was responsible for conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript and financial support.
Corresponding author
Ethics declarations
Conflict of Interest
All authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
All the authors gave consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12033_2021_305_MOESM1_ESM.tif
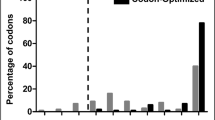
Supplementary file1 (TIF 14273 KB) Fig. S1 Evaluation of human SOX2 protein-coding sequence before (non-optimized) and after (codon-optimized) codon optimization using the GRCA tool. The graph shows the percentage distribution of codons that belong to a specific quality group. Codons with the most frequently used codon for a given amino acid in E. coli is assigned a value of 100. The codons which are distributed in codon quality groups having value ≤ 30 are used less frequently by that organism (threshold is shown as a dotted line) and are more likely to impede the expression. The presence of grey bars (i.e. non-optimized) with values ≤ 30 in the graph signified that it had codons that could affect its expression in E. coli. The absence of black bars (i.e. codon-optimized) with values ≤ 30 in the graph signified that it lacked codons that could affect its expression in E. coli.
12033_2021_305_MOESM2_ESM.tif
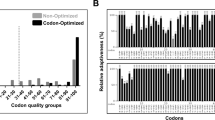
Supplementary file2 (TIF 2228 KB) Fig. S2 Evaluation of human SOX2 protein-coding sequence before (non-optimized) and after (codon-optimized) codon optimization using the GCUA 2.0 tool. The graph shows the codons of non-optimized and codon-optimized SOX2 coding sequence against the relative adaptiveness value. The relative adaptiveness value is based on the number of codons available concerning the organism for that particular amino acid. Therefore, for a given codon, relative adaptiveness value is determined by computing the multiplication of 100 with its usage frequency to the frequently used codon usage frequency. Codons with a relative adaptiveness value ≤ 30% (shown by arrows in black) are very likely to impede the expression in E. coli. The absence of grey bars in the codon-optimized graph shows that it was devoid of inhibitory codons.
Rights and permissions
About this article
Cite this article
Thool, M., Dey, C., Bhattacharyya, S. et al. Generation of a Recombinant Stem Cell-Specific Human SOX2 Protein from Escherichia coli Under Native Conditions. Mol Biotechnol 63, 327–338 (2021). https://doi.org/10.1007/s12033-021-00305-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-021-00305-y