Abstract
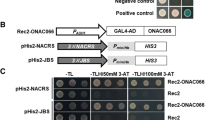
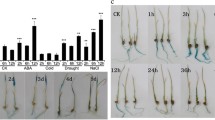
The plant-specific NAC (NAM, ATAF, and CUC)-domain proteins play important roles in plant development and stress responses. In this research, a full-length cDNA named ENAC1 (early NAC-domain protein induced by abiotic stress 1) was isolated from rice. ENAC1 possess one NAC domain in the N-terminus. Comparative time-course expression analysis indicated that ENAC1 expression, similar with OsDREB1A, was induced very quickly by various abiotic stresses including salt, drought, cold, and exogenous abscisic acid. However, the induction of ENAC1 by abiotic stress was transient and lasted up to 3 h, whereas that of OsDREB1A maintained longer. The promoter sequence of ENAC1 harbors several cis-elements including ABA response elements, but the well-known dehydration responsive element/C-repeat element is absent. The ENAC1-GFP (green fluorescent protein) fusion protein was localized in the nucleus of rice protoplast cell. Yeast hybrid assays revealed that ENAC1 was a transcription activator and bound to NAC recognition sequence (NACRS). Co-expression analysis suggested that ENAC1 co-expressed with a number of stress-related genes. Taken together, ENAC1 may be an early transcription activator of stress responses and function in the regulation of NACRS-mediated gene expression under abiotic stress.







Similar content being viewed by others
Abbreviations
- ABA:
-
Abscisic acid
- ABRE:
-
ABA response elements
- CRT:
-
C-repeat
- DAPI:
-
4,6-Diamidino-2-phenylindole
- DRE:
-
Dehydration responsive element
- DREB:
-
Dehydration responsive element binding protein
- GFP:
-
Green fluorescent protein
- NAC:
-
NAM ATAF1/2 CUC2
- NACRS:
-
NAC recognition sequence
- ORF:
-
Open reading frame
- RT:
-
Reverse transcription
References
Olsen, A. N., Ernst, H. A., Leggio, L. L., & Skriver, K. (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends in Plant Science, 10, 79–87.
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., Langum, T. J., Smidt, L., Boomsma, D. D., Emme, N. J., Chen, X., Finer, J. J., Shen, Q. J., & Rushton, P. J. (2011). WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnology Journal. doi:10.1111/j.1467-7652.2001.00634.x.
Huang, J., Sun, S. J., Xu, D. Q., Yang, X., Bao, Y. M., Wang, Z. F., et al. (2009). Increased tolerance of rice to cold, drought and oxidative stresses mediated by the overexpression of a gene that encodes the zinc finger protein ZFP245. Biochemical and Biophysical Research Communications, 389, 556–561.
Huang, J., Wang, M. M., Jiang, Y., Bao, Y. M., Huang, X., Sun, H., et al. (2008). Expression analysis of rice A20/AN1-type zinc finger genes and characterization of ZFP177 that contributes to temperature stress tolerance. Gene, 420, 135–144.
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annual Review of Plant Biology, 53, 247–273.
Kusano, H., Asano, T., Shimada, H., & Kadowaki, K. (2005). Molecular characterization of ONAC300, a novel NAC gene specifically expressed at early stages in various developing tissues of rice. Molecular Genetics and Genomics, 272, 616–626.
Puranik, S., Bahadur, R. P., Srivastava, P. S., & Prasad, M. (2011). Molecular cloning and characterization of a membrane associated NAC family gene, SiNAC from foxtail millet [Setaria italica (L.) P. Beauv.]. Molecular Biotechnology. doi:10.1007/s12033-011-9385-7.
Liu, X., Hong, L., Li, X. Y., Yao, Y., Hu, B., & Li, L. (2011). Improved drought and salt tolerance in transgenic Arabidopsis overexpressing a NAC transcriptional factor from Arachis hypogaea. Bioscience, Biotechnology, and Biochemistry, 75, 443–450.
Tran, L. S., Quach, T. N., Guttikonda, S. K., Aldrich, D. L., Kumar, R., Neelakandan, A., et al. (2009). Molecular characterization of stress-inducible GmNAC genes in soybean. Molecular Genetics and Genomics, 281, 647–664.
Jensen, M. K., Kjaersgaard, T., Nielsen, M. M., Galberg, P., Petersen, K., O’Shea, C., et al. (2010). The Arabidopsis thaliana NAC transcription factor family: Structure–function relationships and determinants of ANAC019 stress signalling. Biochemical Journal, 426, 183–196.
Nuruzzaman, M., Manimekalai, R., Sharoni, A. M., Satoh, K., Kondoh, H., Ooka, H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene, 465, 30–44.
Pinheiro, G. L., Marques, C. S., Costa, M. D., Reis, P. A., Alves, M. S., Carvalho, C. M., et al. (2009). Complete inventory of soybean NAC transcription factors: Sequence conservation and expression analysis uncover their distinct roles in stress response. Gene, 444, 10–23.
Hu, H., Dai, M., Yao, J., Xiao, B., Li, X., Zhang, Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences of the USA, 103, 12987–12992.
Hu, H., You, J., Fang, Y., Zhu, X., Qi, Z., & Xiong, L. (2008). Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Molecular Biology, 67, 169–181.
Yamaguchi-Shinozaki, K., & Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell, 6, 251–264.
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., & Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proceedings of the National Academy of Sciences of the USA, 97, 11632–11637.
Tran, L. S., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell, 16, 2481–2498.
Sun, S. J., Guo, S. Q., Yang, X., Bao, Y. M., Tang, H. J., Sun, H., et al. (2010). Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. Journal of Experimental Botany, 61, 2807–2818.
Reece, K. S., McElroy, D., & Wu, R. (1990). Genomic nucleotide sequence of four rice (Oryza sativa) actin genes. Plant Molecular Biology, 14, 621–624.
Jung, K. H., Dardick, C., Bartley, L. E., Cao, P., Phetsom, J., Canlas, P., et al. (2008). Refinement of light-responsive transcript lists using rice oligonucleotide arrays: Evaluation of gene-redundancy. PLoS One, 3, e3337.
Fang, Y., You, J., Xie, K., Xie, W., & Xiong, L. (2008). Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Molecular Genetics and Genomics, 280, 547–563.
He, X. J., Mu, R. L., Cao, W. H., Zhang, Z. G., Zhang, J. S., & Chen, S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant Journal, 44, 903–916.
Ohnishi, T., Sugahara, S., Yamada, T., Kikuchi, K., Yoshiba, Y., Hirano, H. Y., et al. (2005). OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes and Genetic Systems, 80, 135–139.
Takasaki, H., Maruyama, K., Kidokoro, S., Ito, Y., Fujita, Y., Shinozaki, K., et al. (2010). The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Molecular Genetics and Genomics, 284, 173–183.
Zheng, X., Chen, B., Lu, G., & Han, B. (2009). Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochemical and Biophysical Research Communications, 379, 985–989.
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., & Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell, 15, 63–78.
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., & Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnology, 17, 287–291.
Hao, Y. J., Wei, W., Song, Q. X., Chen, H. W., Zhang, Y. Q., Wang, F., Zou, H. F., Lei, G., Tian, A. G., Zhang, W. K., Ma, B., Zhang, J. S., & Chen, S. Y. (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant Journal, 68, 302–313.
Kim, S. G., Kim, S. Y., & Park, C. M. (2007). A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta, 226, 647–654.
Kim, S. G., Lee, S., Seo, P. J., Kim, S. K., Kim, J. K., & Park, C. M. (2010). Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics, 95, 56–65.
Gonzalez-Lamothe, R., Tsitsigiannis, D. I., Ludwig, A. A., Panicot, M., Shirasu, K., & Jones, J. D. (2006). The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell, 18, 1067–1083.
Dong, C. H., Agarwal, M., Zhang, Y., Xie, Q., & Zhu, J. K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proceedings of the National Academy of Sciences of the USA, 103, 8281–8286.
Zhang, Y., Yang, C., Li, Y., Zheng, N., Chen, H., Zhao, Q., et al. (2007). SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell, 19, 1912–1929.
Gao, T., Wu, Y., Zhang, Y., Liu, L., Ning, Y., Wang, D., et al. (2011). OsSDIR1 overexpression greatly improves drought tolerance in transgenic rice. Plant Molecular Biology, 76, 145–156.
Bos, J. I., Armstrong, M. R., Gilroy, E. M., Boevink, P. C., Hein, I., Taylor, R. M., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proceedings of the National Academy of Sciences of the USA, 107, 9909–9914.
Shimono, M., Sugano, S., Nakayama, A., Jiang, C. J., Ono, K., Toki, S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell, 19, 2064–2076.
Shimono, M., Koga, H., Akagi, A., Hayashi, N., Goto, S., Sawada, M., Kurihara, T., Matsushita, A., Sugano, S., Jiang, C. J., Kaku, H., Inoue, H., & Takatsuji, H. (2011). Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Molecular Plant Pathology. doi:10.1111/j.1364-3703.2011.00732.x.
Acknowledgements
This work was supported by the Program for New Century Excellent Talents in University (no. NCET-08-0795), the National Natural Science Foundation of China (nos 30971556, 30971758), the Research Fund for the Doctoral Program of Higher Education (no. 200803070036), State Key Laboratory of Rice Biology (no. 110101) and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Sun, H., Huang, X., Xu, X. et al. ENAC1, a NAC Transcription Factor, is an Early and Transient Response Regulator Induced by Abiotic Stress in Rice (Oryza sativa L.). Mol Biotechnol 52, 101–110 (2012). https://doi.org/10.1007/s12033-011-9477-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-011-9477-4