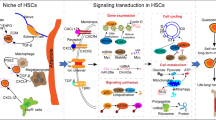
Abstract
Blood is generated throughout life by continued proliferation and differentiation of hematopoietic progenitors, while at the top of the hierarchy, hematopoietic stem cells (HSCs) remain largely quiescent. This way HSCs avoid senescence and preserve their capacity to repopulate the hematopoietic system. But HSCs are not always quiescent, proliferating extensively in conditions such as those found in the fetal liver. Understanding the elusive mechanisms that regulate HSC fate would enable us to comprehend a crucial piece of HSC biology and pave the way for ex-vivo HSC expansion with clear clinical benefit. Here we review how metabolism, endoplasmic reticulum stress and oxidative stress condition impact HSCs decision to self-renew or differentiate and how these signals integrate into the mammalian target of rapamycin (mTOR) pathway. We argue that the bone marrow microenvironment continuously favors differentiation through the activation of the mTOR complex (mTORC)1 signaling, while the fetal liver microenvironment favors self-renewal through the inverse mechanism. In addition, we also postulate that strategies that have successfully achieved HSC expansion, directly or indirectly, lead to the inactivation of mTORC1. Finally, we propose a mechanism by which mTOR signaling, during cell division, conditions HSC fate. This mechanism has already been demonstrated in mature hematopoietic cells (T-cells), that face a similar decision after activation, either undergoing clonal expansion or differentiation.
Graphical Abstract




Similar content being viewed by others
Data Availability
Not applicable.
References
Medvinsky, A., et al. (1996). Definitive hematopoiesis is autonomously initiated by the AGM region. Cell, 86(6), 897–906.
Ema, H., et al. (2000). Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood, 95(7), 2284–2288.
Ueda, T., et al. (2019). Endothelial Cell-Selective Adhesion Molecule Contributes to the Development of Definitive Hematopoiesis in the Fetal Liver. Stem Cell Reports, 13(6), 992–1005.
Khan, J. A., et al. (2016). Fetal liver hematopoietic stem cell niches associate with portal vessels. Science, 351(6269), 176–180.
Chou, S., et al. (2013) Fetal hepatic progenitors support long-term expansion of hematopoietic stem cells. Experimental Hematology, 41(5), 479–490 e4.
Wolber, F. M., et al. (2002). Roles of spleen and liver in development of the murine hematopoietic system. Experimental Hematology, 30(9), 1010–1019.
Christensen, J. L., et al. (2004). Circulation and chemotaxis of fetal hematopoietic stem cells. PLoS Biology, 2(3), E75.
Birbrair, A., et al. (2016). Niche heterogeneity in the bone marrow. Annals of the New York Academy of Sciences, 1370(1), 82–96.
Ditadi, A., et al. (2017). A view of human haematopoietic development from the Petri dish. Nature Reviews. Molecular Cell Biology, 18(1), 56–67.
Pinho, S., et al. (2019). Haematopoietic stem cell activity and interactions with the niche. Nature Reviews. Molecular Cell Biology, 20(5), 303–320.
Rosental, B., et al. (2018). Complex mammalian-like haematopoietic system found in a colonial chordate. Nature, 564(7736), 425–429.
Bowie, M. B., et al. (2006). Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. The Journal of Clinical Investigation, 116(10), 2808–2816.
Wilson, A., et al. (2008). Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell, 135(6), 1118–1129.
Jiang, L., et al. (2018). SHP-1 regulates hematopoietic stem cell quiescence by coordinating TGF-beta signaling. The Journal of Experimental Medicine, 215(5), 1337–1347.
Hirata, Y., et al. (2018). CD150(high) bone marrow tregs maintain hematopoietic stem cell quiescence and immune privilege via adenosine. Cell Stem Cell, 22(3), 445-453 e5.
Fleischman, R. A., et al. (1984). Development of adult bone marrow stem cells in H-2-compatible and -incompatible mouse fetuses. The Journal of Experimental Medicine, 159(3), 731–745.
Papa, L., et al. (2020). Ex vivo HSC expansion challenges the paradigm of unidirectional human hematopoiesis. Annals of the New York Academy of Sciences, 1466(1), 39–50.
Wilkinson, A. C., et al. (2020). Long-term ex vivo expansion of mouse hematopoietic stem cells. Nature Protocols, 15(2), 628–648.
Papadopoli, D., Boulay, K., Kazak, L., Pollak, M., Mallette, F., Topisirovic, I., et al. (2019). mTOR as a central regulator of lifespan and aging, F1000Res, 8. https://doi.org/10.12688/f1000research.17196.1.
Ersahin, T., et al. (2015). The PI3K/AKT/mTOR interactive pathway. Molecular BioSystems, 11(7), 1946–1954.
Jhanwar-Uniyal, M., et al. (2019). Diverse signaling mechanisms of mTOR complexes: mTORC1 and mTORC2 in forming a formidable relationship. Advances in Biological Regulation, 72, 51–62.
Ben-Sahra, I., et al. (2017). mTORC1 signaling and the metabolic control of cell growth. Current Opinion in Cell Biology, 45, 72–82.
Su, K. H., & Dai, C. (2017). mTORC1 senses stresses: Coupling stress to proteostasis, Bioessays, 39(5). https://doi.org/10.12688/f1000research.17196.1.
Rajesh, K., et al. (2015). Phosphorylation of the translation initiation factor eIF2alpha at serine 51 determines the cell fate decisions of Akt in response to oxidative stress. Cell Death & Disease, 6, e1591.
Morita, M., et al. (2015). mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle, 14(4), 473–480.
Dunlop, E. A., et al. (2014). mTOR and autophagy: a dynamic relationship governed by nutrients and energy. Seminars in Cell & Developmental Biology, 36, 121–129.
Lee, G., et al. (2017). Post-transcriptional Regulation of De Novo Lipogenesis by mTORC1-S6K1-SRPK2 Signaling. Cell, 171(7), 1545–1558 e18.
Ben-Sahra, I., et al. (2016). mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science, 351(6274), 728–733.
Khan, N. A., et al. (2017). mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metabolism, 26(2), 419–428.
Xu, W., et al. (2019). CD146 regulates growth factor-induced mTORC2 activity independent of the PI3K and mTORC1 pathways. Cell Reports, 29(5), 1311-1322 e5.
Ebner, M., et al. (2017). Localization of mTORC2 activity inside cells. The Journal of Cell Biology, 216(2), 343–353.
Sen, B., et al. (2014). mTORC2 regulates mechanically induced cytoskeletal reorganization and lineage selection in marrow-derived mesenchymal stem cells. Journal of Bone and Mineral Research, 29(1), 78–89.
Devreotes, P., et al. (2015). Signaling networks that regulate cell migration. Cold Spring Harbor Perspectives in Biology, 7(8), a005959.
Lee, S. E., et al. (2012). mTOR is required for asymmetric division through small GTPases in mouse oocytes. Molecular Reproduction & Development, 79(5), 356–366.
Zou, Z., et al. (2015). mTORC2 promotes cell survival through c-Myc-dependent up-regulation of E2F1. The Journal of Cell Biology, 211(1), 105–122.
Martinez Calejman, C., et al. (2020). mTORC2-AKT signaling to ATP-citrate lyase drives brown adipogenesis and de novo lipogenesis. Nature Communications, 11(1), 575.
Dan, H. C., et al. (2016). PI3K/Akt promotes feedforward mTORC2 activation through IKKalpha. Oncotarget, 7(16), 21064–21075.
Vadysirisack, D. D., et al. (2012). mTOR activity under hypoxia. Methods in Molecular Biology, 821, 45–58.
Wolff, N. C., et al. (2011). Cell-type-dependent regulation of mTORC1 by REDD1 and the tumor suppressors TSC1/TSC2 and LKB1 in response to hypoxia. Molecular and Cellular Biology, 31(9), 1870–1884.
Cam, H., et al. (2010). mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1alpha. Molecular Cell, 40(4), 509–520.
Yan, T., et al. (2017). Hypoxia regulates mTORC1-mediated keratinocyte motility and migration via the AMPK pathway. PLoS One, 12(1), e0169155.
Zhang, J., et al. (2019). Involvement of autophagy in hypoxia-BNIP3 signaling to promote epidermal keratinocyte migration. Cell Death & Disease, 10(3), 234.
Cam, H., et al. (2011). Regulation of mammalian target of rapamycin complex 1 (mTORC1) by hypoxia: causes and consequences. Targeted Oncology, 6(2), 95–102.
Land, S. C., et al. (2007). Hypoxia-inducible factor 1alpha is regulated by the mammalian target of rapamycin (mTOR) via an mTOR signaling motif. The Journal of Biological Chemistry, 282(28), 20534–20543.
Kunisaki, Y., et al. (2013). Arteriolar niches maintain haematopoietic stem cell quiescence. Nature, 502(7473), 637–643.
Yamazaki, S., et al. (2011). Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell, 147(5), 1146–1158.
Bruns, I., et al. (2014). Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nature Medicine, 20(11), 1315–1320.
Tesio, M., et al. (2015). Hematopoietic stem cell quiescence and function are controlled by the CYLD-TRAF2-p38MAPK pathway. The Journal of Experimental Medicine, 212(4), 525–538.
Nakagawa, M. M., et al. (2018). Constitutive activation of NF-kappaB pathway in hematopoietic stem cells causes loss of quiescence and deregulated transcription factor networks. Frontiers in Cell and Development Biology, 6, 143.
Liang, R., Arif, T., Kalmykova, S., Kasianov, A., Lin, M., Menon, V., et al. (2020). Restraining lysosomal activity preserves hematopoietic stem cell quiescence and potency. Cell Stem Cell, 26(3), 359–376.e7. https://doi.org/10.1016/j.stem.2020.01.013.
Zhang, J., et al. (2012). Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell, 11(5), 589–595.
Pei, W., et al. (2017). Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature, 548(7668), 456–460.
Guo, G., et al. (2013). Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell, 13(4), 492–505.
Wu, M., et al. (2007). Imaging hematopoietic precursor division in real time. Cell Stem Cell, 1(5), 541–554.
Morrison, S. J., et al. (1997). Identification of a lineage of multipotent hematopoietic progenitors. Development, 124(10), 1929–1939.
Karamitros, D., et al. (2018). Single-cell analysis reveals the continuum of human lympho-myeloid progenitor cells. Nat Immunol, 19(1), 85–97.
van Galen, P., et al. (2014). Reduced lymphoid lineage priming promotes human hematopoietic stem cell expansion. Cell Stem Cell, 14(1), 94–106.
Cheng, Y., et al. (2019). m(6)A RNA methylation maintains hematopoietic stem cell identity and symmetric commitment. Cell Reports, 28(7), 1703-1716 e6.
Till, J. E., et al. (1961). A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiation Research, 14, 213–222.
Busch, K., et al. (2015). Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature, 518(7540), 542–546.
Sun, J., et al. (2014). Clonal dynamics of native haematopoiesis. Nature, 514(7522), 322–327.
Sawai, C. M., et al. (2016). Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity, 45(3), 597–609.
Rodriguez-Fraticelli, A. E., et al. (2018). Clonal analysis of lineage fate in native haematopoiesis. Nature, 553(7687), 212–216.
Pellin, D., et al. (2019). A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nature Communications, 10(1), 2395.
Carrelha, J., et al. (2018). Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature, 554(7690), 106–111.
Baumgartner, C., et al. (2018). An ERK-dependent feedback mechanism prevents hematopoietic stem cell exhaustion. Cell Stem Cell, 22(6), 879-892 e6.
Regateiro, F. S., et al. (2013). CD73 and adenosine generation in the creation of regulatory microenvironments. Clinical and Experimental Immunology, 171(1), 1–7.
Tusi, B. K., et al. (2018). Population snapshots predict early haematopoietic and erythroid hierarchies. Nature, 555(7694), 54–60.
Hoshii, T., et al. (2014). Pleiotropic roles of mTOR complexes in haemato-lymphopoiesis and leukemogenesis. Journal of Biochemistry, 156(2), 73–83.
Kalaitzidis, D., et al. (2012). mTOR complex 1 plays critical roles in hematopoiesis and Pten-loss-evoked leukemogenesis. Cell Stem Cell, 11(3), 429–439.
Hammerman, P. S., et al. (2005). Pim and Akt oncogenes are independent regulators of hematopoietic cell growth and survival. Blood, 105(11), 4477–4483.
Spevak, C. C., et al. (2020). Hematopoietic stem and progenitor cells exhibit stage-specific translational programs via mTOR- and CDK1-dependent mechanisms. Cell Stem Cell, 26(5), 755-765 e7.
Magee, J. A., et al. (2012). Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell, 11(3), 415–428.
Notario, L., et al. (2018). Anti-CD69 therapy induces rapid mobilization and high proliferation of HSPCs through S1P and mTOR. Leukemia, 32(6), 1445–1457.
Tan, D. Q., et al. (2019). PRMT5 modulates splicing for genome integrity and preserves proteostasis of hematopoietic stem cells. Cell Reports, 26(9), 2316-2328 e6.
Wang, X., et al. (2019). Rheb1 loss leads to increased hematopoietic stem cell proliferation and myeloid-biased differentiation in vivo. Haematologica, 104(2), 245–255.
Peng, H., et al. (2018). Distinct roles of Rheb and Raptor in activating mTOR complex 1 for the self-renewal of hematopoietic stem cells. Biochemical and Biophysical Research Communications, 495(1), 1129–1135.
Buechler, M. B., et al. (2016). Cutting edge: direct sensing of TLR7 ligands and type I IFN by the common myeloid progenitor promotes mTOR/PI3K-dependent emergency myelopoiesis. Journal of Immunology, 197(7), 2577–2582.
Buechler, M. B., et al. (2013). Cutting edge: Type I IFN drives emergency myelopoiesis and peripheral myeloid expansion during chronic TLR7 signaling. Journal of Immunology, 190(3), 886–891.
Smieszek, A., Marcinkowska, K., Pielok, A., Sikora, M., Valihrach, L., & Marycz, K. (2020). The role of miR-21 in osteoblasts-osteoclasts coupling in vitro. Cells, 9(2), 479. https://doi.org/10.3390/cells9020479.
Chen, X., et al. (2013). microRNAs are ligands of Toll-like receptors. RNA, 19(6), 737–739.
Wang, W. L., et al. (2019). Role of rictor in hematopoietic stem cells during fetal liver hematopoiesis. Zhongguo Shi Yan Xue Ye Xue Za Zhi, 27(2), 600–605.
Chen, C., et al. (2008). TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. The Journal of Experimental Medicine, 205(10), 2397–2408.
Weichhart, T. (2018). mTOR as regulator of lifespan, aging, and cellular senescence: a mini-review. Gerontology, 64(2), 127–134.
Wilkinson, J. E., et al. (2012). Rapamycin slows aging in mice. Aging Cell, 11(4), 675–682.
Lopez-Otin, C., et al. (2013). The hallmarks of aging. Cell, 153(6), 1194–1217.
de Haan, G., et al. (2018). Aging of hematopoietic stem cells. Blood, 131(5), 479–487.
Chen, C., et al. (2009). mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Science Signaling, 2, ra75.
Chen, Y. H., et al. (2018). Asymmetric PI3K activity in lymphocytes organized by a PI3K-mediated polarity pathway. Cell Reports, 22(4), 860–868.
Sharif, O., et al. (2019) Macrophage rewiring by nutrient associated PI3K dependent pathways. Frontiers in Immunology, 10, 2002.
Wu, X. L., et al. (2018). Effects of Glut1 gene silencing on proliferation, differentiation, and apoptosis of colorectal cancer cells by targeting the TGF-beta/PI3K-AKT-mTOR signaling pathway. Journal of Cellular Biochemistry, 119(2), 2356–2367.
Buller, C. L., et al. (2008). A GSK-3/TSC2/mTOR pathway regulates glucose uptake and GLUT1 glucose transporter expression. American Journal of Physiology. Cell Physiology, 295(3), C836–C843.
Oburoglu, L., et al. (2014) Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell, 15(2), 169 – 84.
Sarrazy, V., et al. (2016). Disruption of Glut1 in hematopoietic stem cells prevents myelopoiesis and enhanced glucose flux in atheromatous plaques of ApoE(-/-) mice. Circulation Research, 118(7), 1062–1077.
Meugnier, E., et al. (2007). Regulation of gene expression by glucose. Current Opinion in Clinical Nutrition and Metabolic Care, 10(4), 518–522.
Albert, B., et al. (2019). Sfp1 regulates transcriptional networks driving cell growth and division through multiple promoter-binding modes. Genes & Development, 33(5–6), 288–293.
Bond, M. R., et al. (2015). A little sugar goes a long way: the cell biology of O-GlcNAc. The Journal of Cell Biology, 208(7), 869–880.
Saki, N., et al. (2013). Adverse effect of high glucose concentration on stem cell therapy. International Journal of Hematology-Oncology and Stem Cell Research, 7(3), 34–40.
Freund, P., et al. (2017). O-GlcNAcylation of STAT5 controls tyrosine phosphorylation and oncogenic transcription in STAT5-dependent malignancies. Leukemia, 31(10), 2132–2142.
Rauth, M., Freund, P., Orlova, A., Grünert, S., Tasic, N., Han, X., et al. (2019). Cell metabolism control through O-GlcNAcylation of STAT5: A full or empty fuel tank makes a big difference for cancer cell growth and survival. International Journal of Molecular Sciences, 20(5), 1028. https://doi.org/10.3390/ijms20051028.
Dey-Guha, I., et al. (2011). Asymmetric cancer cell division regulated by AKT. Proceedings of the National Academy of Sciences of the United States of America, 108(31), 12845–12850.
Murray, E. R., et al. (2017). Towards specific inhibition of mTORC2. Aging (Albany NY), 9(12), 2461–2462.
Yu, J. S., et al. (2016). Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development, 143(17), 3050–3060.
Murakami, M., et al. (2004). mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Molecular and Cellular Biology, 24(15), 6710–6718.
Shiota, C., et al. (2006). Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Developmental Cell, 11(4), 583–589.
Xu, K., et al. (2014). mTOR signaling in tumorigenesis. Biochimica et Biophysica Acta, 1846(2), 638–654.
Murugan, A. K., et al. (2013). Mutations in critical domains confer the human mTOR gene strong tumorigenicity. The Journal of Biological Chemistry, 288(9), 6511–6521.
Nazareth, E. J. P., et al. (2016). A multi-lineage screen reveals mTORC1 inhibition enhances human pluripotent stem cell mesendoderm and blood progenitor production. Stem Cell Reports, 6(5), 679–691.
Mohammad, K., Dakik, P., Medkour, Y., Mitrofanova, D., & Titorenko, V. I. (2019). Quiescence entry, maintenance, and exit in adult stem cells. International Journal of Molecular Sciences, 20(9), 2158. https://doi.org/10.3390/ijms20092158.
LiCausi, F., & Hartman, N. W. (2018). Role of mTOR complexes in neurogenesis. International Journal of Molecular Sciences, 19(5), 1544. https://doi.org/10.3390/ijms19051544.
Castilho, R. M., et al. (2009). mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell, 5(3), 279–289.
Deng, Z., et al. (2015). mTOR signaling promotes stem cell activation via counterbalancing BMP-mediated suppression during hair regeneration. Journal of Molecular Cell Biology, 7(1), 62–72.
Rion, N., Castets, P., Lin, S., Enderle, L., Reinhard, J. R., Eickhorst, C., et al. (2019). mTOR controls embryonic and adult myogenesis via mTORC1. Development, 146(7). https://doi.org/10.1242/dev.172460.
Moreira, B. P., Oliveira, P. F., & Alves, M. G. (2019). Molecular mechanisms controlled by mTOR in male reproductive system. International Journal of Molecular Sciences, 20(7), 1633. https://doi.org/10.3390/ijms20071633.
Kaur, H., et al. (2019). Role of mTORC1 in intestinal epithelial repair and tumorigenesis. Cellular and Molecular Life Sciences, 76(13), 2525–2546.
Yousefi, M., et al. (2018). Calorie restriction governs intestinal epithelial regeneration through cell-autonomous regulation of mTORC1 in reserve stem cells. Stem Cell Reports, 10(3), 703–711.
Castilho, R. M., Squarize, C. H., & Gutkind, J. S. (2013). Exploiting PI3K/mTOR signaling to accelerate epithelial wound healing. Oral Diseases, 19(6), 551–558. https://doi.org/10.1111/odi.12070.
Liu, L., et al. (2017). Exosomes derived from mesenchymal stem cells rescue myocardial ischaemia/reperfusion injury by inducing cardiomyocyte autophagy via AMPK and Akt pathways. Cellular Physiology and Biochemistry, 43(1), 52–68.
Hartman, N. W., et al. (2013). mTORC1 targets the translational repressor 4E-BP2, but not S6 kinase 1/2, to regulate neural stem cell self-renewal in vivo. Cell Reports, 5(2), 433–444.
Tan, V. P., et al. (2016). Nutrient-sensing mTORC1: Integration of metabolic and autophagic signals. Journal of Molecular and Cellular Cardiology, 95, 31–41.
Kim, J., et al. (2019). mTOR as a central hub of nutrient signalling and cell growth. Nature Cell Biology, 21(1), 63–71.
Manesia, J. K., et al. (2015). Highly proliferative primitive fetal liver hematopoietic stem cells are fueled by oxidative metabolic pathways. Stem Cell Research, 15(3), 715–721.
Takubo, K., et al. (2013). Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell, 12(1), 49–61.
Guo, B., et al. (2018). Antagonism of PPAR-gamma signaling expands human hematopoietic stem and progenitor cells by enhancing glycolysis. Nature Medicine, 24(3), 360–367.
Bonora, M., et al. (2018). Membrane-potential compensation reveals mitochondrial volume expansion during HSC commitment. Experimental Hematology, 68, 30-37 e1.
de Almeida, M. J., et al. (2017). Dye-independent methods reveal elevated mitochondrial mass in hematopoietic stem cells. Cell Stem Cell, 21(6), 725–729.
Papa, L., et al. (2019). Mitochondrial role in stemness and differentiation of hematopoietic stem cells. Stem Cells International, 2019, 4067162.
Nakamura-Ishizu, A., et al. (2020). Hematopoietic stem cell metabolism during development and aging. Developmental Cell, 54(2), 239–255.
Juntilla, M. M., et al. (2010). AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood, 115(20), 4030–4038.
Khatri, R., et al. (2016). Reactive oxygen species limit the ability of bone marrow stromal cells to support hematopoietic reconstitution in aging mice. Stem Cells and Development, 25(12), 948–958.
Ludin, A., et al. (2014). Reactive oxygen species regulate hematopoietic stem cell self-renewal, migration and development, as well as their bone marrow microenvironment. Antioxidants & Redox Signaling, 21(11), 1605–1619.
Takubo, K., et al. (2010). Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell, 7(3), 391–402.
Kajla, S., et al. (2012). A crucial role for Nox 1 in redox-dependent regulation of Wnt-beta-catenin signaling. The FASEB Journal, 26(5), 2049–2059.
Duncan, A. W., et al. (2005). Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nature Immunology, 6(3), 314–322.
Itoh, M., et al. (2019). Hypoxia Up-regulates HIF expression while suppressing cell growth and NOTCH activity in leukaemia cells. Anticancer Research, 39(8), 4165–4170.
Miharada, K., et al. (2014). Dppa5 improves hematopoietic stem cell activity by reducing endoplasmic reticulum stress. Cell Reports, 7(5), 1381–1392.
Sigurdsson, V., et al. (2016). Bile acids protect expanding hematopoietic stem cells from unfolded protein stress in fetal liver. Cell Stem Cell, 18(4), 522–532.
Crawford, L. W., et al. (2010). Histology atlas of the developing mouse hepatobiliary system with emphasis on embryonic days 9.5–18.5. Toxicologic Pathology, 38(6), 872–906.
Reimold, A. M., et al. (2000). An essential role in liver development for transcription factor XBP-1. Genes & Development, 14(2), 152–157.
Zhang, C. C., et al. (2014) Hypoxia and metabolic properties of hematopoietic stem cells, Antioxidants & Redox Signaling, 20(12), 1891 – 901.
Wouters, B. G., et al. (2008). Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nature Reviews. Cancer, 8(11), 851–864.
Delbrel, E., et al. (2018). HIF-1alpha triggers ER stress and CHOP-mediated apoptosis in alveolar epithelial cells, a key event in pulmonary fibrosis. Scientific Reports, 8(1), 17939.
Song, S., et al. (2018). Intermittent-hypoxia-induced autophagy activation through the ER-stress-related PERK/eIF2alpha/ATF4 pathway is a protective response to pancreatic beta-cell apoptosis. Cellular Physiology and Biochemistry, 51(6), 2955–2971.
Bettigole, S. E., et al. (2015). The transcription factor XBP1 is selectively required for eosinophil differentiation. Nature Immunology, 16(8), 829–837.
Zheng, P., et al. (2013). Cytopenia and autoimmune diseases: a vicious cycle fueled by mTOR dysregulation in hematopoietic stem cells. Journal of Autoimmunity, 41, 182–187.
Miriuka, S. G., et al. (2006). mTOR inhibition induces endothelial progenitor cell death. American Journal of Transplantation, 6(9), 2069–2079.
Darici, S., et al. (2020) Targeting PI3K/Akt/mTOR in AML: Rationale and clinical evidence. Journal of Clinical Medicine, 9(9). https://doi.org/10.1111/odi.12070.
Yang, A., et al. (2015). Differential reponses of hematopoietic stem and progenitor cells to mTOR inhibition. Stem Cells International, 2015, 561404.
Luo, Y., et al. (2014). Rapamycin enhances long-term hematopoietic reconstitution of ex vivo expanded mouse hematopoietic stem cells by inhibiting senescence. Transplantation, 97(1), 20–29.
Rohrabaugh, S. L., et al. (2011). Ex vivo rapamycin treatment of human cord blood CD34 + cells enhances their engraftment of NSG mice. Blood Cells, Molecules & Diseases, 46(4), 318–320.
Ito, K., et al. (2004). Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature, 431(7011), 997–1002.
Simsek, T., et al. (2010). The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell, 7(3), 380–390.
Wilkinson, A. C., et al. (2019). Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature, 571(7763), 117–121.
Gadhoum, S. Z., et al. (2016). Anti-CD44 antibodies inhibit both mTORC1 and mTORC2: a new rationale supporting CD44-induced AML differentiation therapy. Leukemia, 30(12), 2397–2401.
Majka, M., et al. (2002). Thrombopoietin, but not cytokines binding to gp130 protein-coupled receptors, activates MAPKp42/44, AKT, and STAT proteins in normal human CD34 + cells, megakaryocytes, and platelets. Experimental Hematology, 30(7), 751–760.
Kunitama, M., et al. (1997). Protein kinase C and c-myc gene activation pathways in thrombopoietin signal transduction. Biochemical and Biophysical Research Communications, 231(2), 290–294.
Chanprasert, S., et al. (2006). Thrombopoietin (TPO) induces c-myc expression through a PI3K- and MAPK-dependent pathway that is not mediated by Akt, PKCzeta or mTOR in TPO-dependent cell lines and primary megakaryocytes. Cellular Signalling, 18(8), 1212–1218.
Satoh, Y., et al. (2004). Roles for c-Myc in self-renewal of hematopoietic stem cells. The Journal of Biological Chemistry, 279(24), 24986–24993.
Bai, T., et al. (2019). Expansion of primitive human hematopoietic stem cells by culture in a zwitterionic hydrogel. Journal of Natural Medicines, 25(10), 1566–1575.
Funding
The authors would like to acknowledge funding from UIDB/04539/2020, Healthy Aging 2020-CENTRO-01-0145-FEDER-000012-N2323, POCI-01-0145-FEDER-007440, and the NIH 5P30 AG028718.
Author information
Authors and Affiliations
Contributions
Hélia Fernandes and João Moura conceived and wrote the manuscript, Eugénia Carvalho critically reviewed and discussed the manuscript. All authors approved the final version of this manuscript.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
Conflict of Interest
The authors have no potential conflicts of interest to disclose.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• The mTOR pathway integrates metabolic, oxidative and endoplasmic reticulum stress and other environmental signals that condition HSC fate.
• Bone marrow microenvironment suppresses HSC expansion through mTOR signaling, protecting HSC from exhaustion.
• Many strategies leading to ex vivo HSC expansion indirectly suppress mTOR signaling.
Rights and permissions
About this article
Cite this article
Fernandes, H., Moura, J. & Carvalho, E. mTOR Signaling as a Regulator of Hematopoietic Stem Cell Fate. Stem Cell Rev and Rep 17, 1312–1322 (2021). https://doi.org/10.1007/s12015-021-10131-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-021-10131-z