Abstract
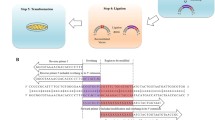
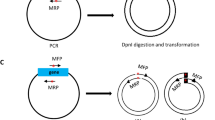
Site-directed mutagenesis is one of the most important tools in molecular biology. The majority of the mutagenesis methods have been developed to mutate one region of target DNA in each cycle of mutagenesis, while in some cases there is a need to mutate several distal points. We used a new method to simultaneously mutate two distal points in the target DNA. Different regions of the target DNA were amplified in three separate PCR reactions. The PCR products were back-to-back and together they made the complete length of the template DNA. Mutations were introduced to PCR products by middle mutagenic primers. PCR products were mixed and ligated with random blunt ligation, and then the desired mutated DNA fragments were selected in two steps by flanking restriction enzyme digestion and size selection. Selected fragments were amplified in another PCR reaction using flanking primers and finally cloned into the plasmid vector. This mutagenesis process is simple, there is no need to use modified primers and long or difficult PCR reactions.




Similar content being viewed by others
References
Boldrin, F., Degiacomi, G., Serafini, A., Kolly, G. S., Ventura, M., Sala, C., Provvedi, R., Palu, G., Cole, S. T., & Manganelli, R. (2018). Promoter mutagenesis for fine-tuning expression of essential genes in mycobacterium tuberculosis. Microbial Biotechnology, 11(1), 238–247.
Pirlot, C., Thiry, M., Trussart, C., Di Valentin, E., Piette, J., & Habraken, Y. (2016). Melanoma antigen-D2: A nucleolar protein undergoing delocalization during cell cycle and after cellular stress. Biochimica et Biophysica Acta, 1863(4), 581–595.
Hsieh, P. C., & Vaisvila, R. (2013). Protein engineering: single or multiple site-directed mutagenesis. Methods in Molecular Biology, 978, 173–186.
Sen, S., Venkata Dasu, V., & Mandal, B. (2007). Developments in directed evolution for improving enzyme functions. Applied Biochemistry and Biotechnology, 143(3), 212–223.
Foster, P. L. (1991). In vivo mutagenesis. Methods in Enzymology, 204, 114–125.
Bose, J. L. (2016). Chemical and UV mutagenesis. Methods in Molecular Biology, 1373, 111–115.
Tan, J., Chu, J., Hao, Y., Guo, Y., Zhuang, Y., & Zhang, S. (2013). High-throughput system for screening of cephalosporin C high-yield strain by 48-deep-well microtiter plates. Applied Biochemistry and Biotechnology, 169(5), 1683–1695.
Hoshijima, K., Jurynec, M. J., & Grunwald, D. J. (2016). Precise genome editing by homologous recombination. Methods in Cell Biology, 135, 121–147.
Ren, C., Liu, X., Zhang, Z., Wang, Y., Duan, W., Li, S., & Liang, Z. (2016). CRISPR/Cas9-mediated efficient targeted mutagenesis in chardonnay (Vitis vinifera L.). Scientific Reports, 6(1), 32289.
Zhang, J., Zhu, Z., Yue, W., Li, J., Chen, Q., Yan, Y., Lei, A., & Hua, J. (2019). Establishment of CRISPR/Cas9-mediated knock-in system for porcine cells with high efficiency. Applied Biochemistry and Biotechnology.
Botstein, D., & Shortle, D. (1985). Strategies and applications of in vitro mutagenesis. Science, 229(4719), 1193–1201.
Ling, M. M., & Robinson, B. H. (1997). Approaches to DNA mutagenesis: an overview. Analytical Biochemistry, 254(2), 157–178.
Reikofski, J., & Tao, B. Y. (1992). Polymerase chain reaction (PCR) techniques for site-directed mutagenesis. Biotechnology Advances, 10(4), 535–547.
Smith, M. (1985). In vitro mutagenesis. Annual Review of Genetics, 19(1), 423–462.
Karimi, E., Karkhane, A. A., Yakhchali, B., Shamsara, M., Aminzadeh, S., Torktaz, I., Hosseini, M., & Safari, Z. (2014). Study of the effect of F17A mutation on characteristics of Bacillus thermocatenulatus lipase expressed in Pichia pastoris using in silico and experimental methods. Biotechnology and Applied Biochemistry, 61(3), 264–273.
Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., & Pease, L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77(1), 51–59.
Hemsley, A., Arnheim, N., Toney, M. D., Cortopassi, G., & Galas, D. J. (1989). A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Research, 17(16), 6545–6551.
Qi, D., & Scholthof, K. B. (2008). A one-step PCR-based method for rapid and efficient site-directed fragment deletion, insertion, and substitution mutagenesis. Journal of Virological Methods, 149(1), 85–90.
Sanchis, J., Fernandez, L., Carballeira, J. D., Drone, J., Gumulya, Y., Hobenreich, H., Kahakeaw, D., Kille, S., Lohmer, R., Peyralans, J. J., Podtetenieff, J., Prasad, S., Soni, P., Taglieber, A., Wu, S., Zilly, F. E., & Reetz, M. T. (2008). Improved PCR method for the creation of saturation mutagenesis libraries in directed evolution: application to difficult-to-amplify templates. Applied Microbiology and Biotechnology, 81(2), 387–397.
Yu, G., Jia, X., Wen, J., Lu, W., Wang, G., Caiyin, Q., & Chen, Y. (2011). Strain improvement of Streptomyces roseosporus for daptomycin production by rational screening of He-Ne laser and NTG induced mutants and kinetic modeling. Applied Biochemistry and Biotechnology, 163(6), 729–743.
Zhang, W., & Mannervik, B. (2013). An improved dual-tube megaprimer approach for multi-site saturation mutagenesis. World Journal of Microbiology and Biotechnology, 29(4), 667–672.
Luo, W.-G., Liu, H.-Z., & Lin, W.-H. (2013). Simultaneous splicing of multiple DNA fragments in one PCR reaction. Biol. proced. online, 15(1), 9.
Tseng, W. C., Lin, J. W., Hung, X. G., & Fang, T. Y. (2010). Simultaneous mutations up to six distal sites using a phosphorylation-free and ligase-free polymerase chain reaction-based mutagenesis. Analytical Biochemistry, 401(2), 315–317.
Fushan, A. A., & Drayna, D. T. (2009). MALS: an efficient strategy for multiple site-directed mutagenesis employing a combination of DNA amplification, ligation and suppression PCR. BMC Biotechnology, 9(1), 83.
Ali, S. A., & Steinkasserer, A. (1995). PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques, 18, 746–749.
Adereth, Y., Champion, K. J., Hsu, T., & Dammai, V. (2005). Site-directed mutagenesis using Pfu DNA polymerase and T4 DNA ligase. BioTechniques, 38(6), 864–868.
Wurch, T., Lestienne, F., & Pauwels, P. J. (1998). A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotechnology Techniques, 12(9), 653–657.
Peng, R.-H., Xiong, A.-S., & Yao, Q.-H. (2006). A direct and efficient PAGE-mediated overlap extension PCR method for gene multiple-site mutagenesis. Applied Microbiology and Biotechnology, 73(1), 234–240.
Mergulhão, F., Kelly, A., Monteiro, G., Taipa, M., & Cabral, J. (1999). Troubleshooting in gene splicing by overlap extension. Molecular Biotechnology, 12(3), 285–287.
Funding
This study was supported by the National Institute for Genetic Engineering and Biotechnology and the University of Sistan and Baluchestan.
Author information
Authors and Affiliations
Contributions
B. Yakhchali, A. A. Karkhane, and M. H. Sangtarash conceived and designed the research. J. Khezri performed experiments. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khezri, J., Yakhchali, B., Karkhane, A.A. et al. An Efficient Approach for Two Distal Point Site-Directed Mutagenesis from Randomly Ligated PCR Products. Appl Biochem Biotechnol 189, 1318–1326 (2019). https://doi.org/10.1007/s12010-019-03059-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-019-03059-1