Abstract
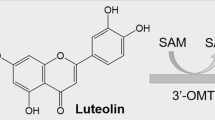
Methylation is a common post-modification reaction that is observed during the biosynthesis of secondary metabolites produced by plants and microorganisms. Based on the sequence information from Streptomyces peucetius ATCC27952, a putative O-methyltransferase (OMT) gene SpOMT7740 was polymerase chain reaction amplified and cloned into E. coli BL21 (DE3) host to test the substrate promiscuity and conduct functional characterization. In vitro and in vivo reaction assays were carried out over various classes of substrates: flavonoids (flavonol, flavones, and isoflavonoid), chalcones, anthraquinones, anthracyclines, and sterol molecules, and the applications in synthesizing diverse classes of O-methoxy natural products were also illustrated. SpOMT7740 catalyzed the O-methylation reaction to form various natural and non-natural O-methoxides, includes 7-hydroxy-8-O-methoxy flavone, 3-O-methoxy flavone, three mono-, di-, and tri-O-methoxy genistein, mono-O-methoxy phloretin, mono-O-methoxy luteolin, 3-O-methoxy β-sitosterol, and O-methoxy anthraquinones (emodin and aloe emodin) and O-methoxy anthracycline (daunorubicin) exhibiting diverse substrate flexibility. Daunorubicin is a native secondary metabolite of S. peucetius. Among the compounds tested, 7,8-dihydroxyflavone was the best substrate for bioconversion to 7-hydroxy-8-O-methoxy flavone, and it was structurally elucidated. This enzyme showed a flexible catalysis over the given ranges of temperature, pH, and divalent cationic conditions for O-methylation.





Similar content being viewed by others
References
Niraula, N. P., Kim, S. H., Sohng, J. K., & Kim, E. S. (2010). Biotechnological doxorubicin production: pathway and regulation engineering of strains for enhanced production. Applied Microbiology and Biotechnology, 87, 1187–1194.
Watve, M. G., Tickoo, R., Jog, M. M., & Bhole, B. D. (2001). How many antibiotics are produced by the genus Streptomyces. Archives of Microbiology, 176, 386–390.
Yoon, Y., Yi, Y. S., Lee, Y., Kim, S., Kim, B. G., Ahn, J. H., & Lim, Y. (2005). Characterization of O-methyltransferase ScOMT1 cloned from Streptomyces coelicolor A3(2). Biochimica et Biophysica Acta, 1730, 85–95.
Choi, K. Y., Jung, E., Yang, Y. H., & Kim, B. G. (2013). Production of a novel O-methyl-isoflavone by regioselective sequential hydroxylation and O-methylation reactions in Streptomyces avermitilis host system. Biotechnology and Bioengineering, 110, 2591–2599.
Olano, C., Mendez, C., & Salas, J. A. (2010). Post-PKS tailoring steps in natural product-producing actinomycetes from the perspective of combinatorial biosynthesis. Natural Product Reports, 27, 571–616.
Martin, J. L., & McMillan, F. M. (2002). SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Current Opinion in Structural Biology, 12, 783–793.
Ibdah, M., Zhang, X. H., Schmidt, J., & Voqt, T. (2003). A novel Mg(2+)-dependent O-methyltransferase in the phenylpropanoid metabolism of mesembryanthemum crystallium. Journal of Biological Chemistry, 278, 43961–43972.
NDong, C., Anzellotti, D., Ibrahim, R. K., Huner, N. P., & Sarhan, F. (2003). Daphnetin methylation by a novel O-methyltransferase is associated with cold acclimation and photosystem II excitation pressure in rye. Journal of Biological Chemistry, 278, 6854–6861.
He, X. Z., Reddy, J. T., & Dixon, R. A. (1998). Stress responses in alfalfa (Medicago sativa L). XXII. cDNA cloning and characterization of an elicitor-inducible isoflavone 7-O-methyltransferase. Plant Molecular Biology, 36, 43–54.
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology and biotechnology. Plant Physiology, 126, 485–493.
Koirala, N., Pandey, R. P., Parajuli, P., Jung, H. J., & Sohng, J. K. (2014). Methylation and subsequent glycosylation of 7,8-dihydroxyflavone. Journal of Biotechnology, 184, 128–137.
Kim, B. G., Jung, B. R., Lee, Y., Hur, H. G., Lim, Y., & Ahn, J. H. (2006). Regiospecific flavonoid 7-O-methylation with Streptomyces avermitilis O-methyltransferase expressed in Escherichia coli. Journal of Agricultural and Food Chemistry, 54, 823–828.
Jnawali, H. N., Lee, E., Jeong, K. W., Shin, A., Heo, Y. S., & Kim, Y. (2014). Anti-inflammatory activity of rhamnetin and a model of its binding to c-Jun NH2-terminal kinase 1 and p38 MAPK. Journal of Natural Products, 77, 258–263.
Hernandez, V., Recio, M. C., Menez, S., Giner, R. M., & Rios, J. L. (2007). Effects of naturally occurring dihydroflavonols from Inula viscosa on inflammation and enzymes involved in the arachidonic acid metabolism. Life Sciences, 81, 480–488.
Zhang, L., Kong, Y., Zhang, H., Wu, H. J., Chen, J., Ding, J., Hu, L., Jiang, H., & Shen, X. (2008). Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobater pylori: Crystal structure characterization with enzymatic inhibition assay. Protein Science, 17, 1971–1978.
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., & Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876–4882.
Dereeper, A., Audic, S., Claverie, J. M., & Blanc, G. (2010). BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evolutionary Biology, 10, 8.
Sambrook, J., & Russell, D. W. (2001). Molecular cloning: a laboratory manual, 3rd ed. NY.
Pandey, R. P., Gurung, R. B., Parajuli, P., Koirala, N., Tuoi Le, T., & Sohng, J. K. (2014). Assessing acceptor substrate promiscuity of YjiC-mediated glycosylation towards flavonoids. Carbohydrate Research, 393, 26–31.
Parajuli, P., Pandey, R. P., Koirala, N., Yoon, Y. J., Kim, B. G., & Sohng, J. K. (2014). Enzymatic synthesis of epothilone A glycosides. AMB Express, 4, 31.
Madduri, K., Torti, F., Colombo, A. L., & Hutchinson, C. R. (1993). Cloning and sequencing of a gene encoding carminomycin 4-O-methyltransferase from Streptomyces peucetius and its expression in E. coli. Journal of Bacteriology, 175, 3900–3904.
Kim, N. Y., Kim, J. H., Lee, Y. H., Lee, E. J., Kim, J., Lim, Y., Chong, Y., & Ahn, J. H. (2007). O-methylation of flavonoids using DnrK based on molecular docking. Journal of Microbiology, 17, 1991–1995.
Grocholski, T., Dinis, P., Niiranen, L., Niemi, J., & Metsa-Ketela, M. (2015). Divergent evolution of a typical S-adenosyl- l -methionine-dependent monooxygenase involved in anthracycline biosynthesis. Proceeding of the National Academy of Sciences of the United States of America, 112, 98866-9871.
Huber, T. D., Wang, F., Songh, S., Johnson, B. R., Zhang, J., Sunkara, M., Van Lanen, S. G., Morris, A. J., Phillips Jr., G. N., & Thorson, J. S. (2016). Functional AdoMet isosteres resistant to classical AdoMet degradation pathways. ACS Chemical Biology, 11, 2484–2491.
Liu, W., Christenson, S. D., Standage, S., & Shen, B. (2002). Biosynthesis of the enedyne antitumor antibiotic C-1027. Science, 297, 1170–1173.
Liu, W., Nonaka, K., Jie, L., Zhang, J., Christenson, S. D., Bae, J., Van Lanen, S. G., Zazopoulos, E., Farnet, C. M., Yang, C. F., & Shen, B. (2005). The neocarzinostatin biosynthetic gene cluster from Streptomyces carzinostaticus ATCC 15944 involving two iterative type I polyketide synthases. Chemical Biology, 12, 293–302.
Chiang, C. M., Ding, H. Y., Tsai, Y. T., & Chang, T. S. (2015). Production of two novel methoxy-isoflavones from biotransformation of 8-hydroxydaidzein by recombinant E. coli expression O-methyltransferase SpOMT2884 from S. peucetius. International Journal of Molecular Sciences, 16, 27816–27823.
Hortobagyi, G. N. (1997). Anthracyclines in the treatment of cancer. Drugs, 4, 1–7.
Rivankar, S. (2014). An overview of doxorubicin formulations in cancer therapy. Journal of Cancer Research Therapeutics, 10, 853–858.
Bansal, G., & Kaushik, D. (2015). Four new degredation products of doxorubicin: an application of forced degradation study and hyphenated chromatographic techniques. Journal of Pharmaceutical Analysis, 5, 285–295.
Parajuli, P., Pandey, R. P., Trang, N. T., Chaudhary, A. K., & Sohng, J. K. (2015). Synthetic sugar cassettes for the efficient production of flavonol glycosides in Escherichia coli. Microbial Cell Factories, 14, 76.
Pandey, R. P., Parajuli, P., Koffas, M. A., & Sohng, J. K. (2016). Microbial production of natural and non-natural flavonoids: pathway engineering, directed evolution and systems/synthetic biology. Biotechnology Advances, 34, 634–662.
Dhar, K., & Rosazza, J. P. (2000). Purification and characterization of Streptomyces griseus catechol O-methyltransferase. Applied and Environmental Microbiology, 66, 4877–4882.
Malla, S., Koffas, M. A., Kazlauskas, R. J., & Kim, B. G. (2012). Production of 7-O-methyl aromadendrin, a medicinally valuable flavonoid, in Escherichia coli. Applied and Environmental Microbiology, 78, 684–694.
Zhang, Y., Han, M. Z., Zhu, S. L., Li, M., Dong, X., Kong, Z., Lu, Y. X., Wang, S. Y., & Tong, W. Y. (2015). Studies on the function and catalytic mechanism of O-methyltransferase SviOMT02, SviOMT03 and SviOMT06 from Streptomyces virginiae IBL14. Enzyme Microbial Technology, 74, 72–79.
Yoon, Y., Park, Y., Lee, Y., Yi, Y. S., Jo, G., Park, J. C., Ahn, J. H., & Lim, Y. (2010). Characterization of an O-methyltransferase from Streptomyces avermitilis MA-4680. Journal of Microbiology and Biotechnology, 20, 1359–1366.
Jansson, A., Koskiniemi, H., Mantsala, P., Niemi, J., & Schneider, G. (2004). Crystal structure of a ternary complex of DnrK, a methyltransferase in daunorubicin biosynthesis, with bound products. Journal of Biological Chemistry, 279, 41149–41156.
Funding Information
This work was supported by a grant from the Next-Generation BioGreen 21 Program (SSAC, grant no.: PJ01111901), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interests.
Electronic supplementary material
ESM 1
(DOCX 1567 kb)
Rights and permissions
About this article
Cite this article
Parajuli, P., Pandey, R.P., Nguyen, T.H.T. et al. Substrate Scope of O-Methyltransferase from Streptomyces peucetius for Biosynthesis of Diverse Natural Products Methoxides. Appl Biochem Biotechnol 184, 1404–1420 (2018). https://doi.org/10.1007/s12010-017-2603-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-017-2603-4