Abstract
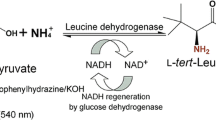
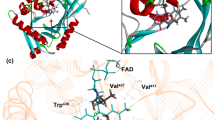
L-2-aminobutyric acid (L-ABA) as a precursor for the anticonvulsant and the antituberculotic is a key intermediate in the chemical and pharmaceutical industries. Recently, leucine dehydrogenase (LeuDH) with NAD+ regeneration was developed for L-ABA production on a large scale. Previously, the L-ABA yield was improved by optimizing conversion conditions, including cofactor regeneration and enzyme immobilization but not protein engineering on LeuDH due to lacking an applicable high-throughput screening (HTS) method. Recently, an HTS assay was developed by us, which enables researchers to engineer LeuDH in a relatively short period of time. Herein, a semirational engineering was performed on LeuDH to increase the catalytic efficiency of BcLeuDH. Firstly, the structure of wild-type (WT) BcLeuDH was modeled and seven potentially beneficial positions were selected for mutation. Five beneficial variants were then identified from the seven site-saturation mutagenesis (SSM) libraries by HTS and confirmed by rescreening via amino acid analyzer. The “best” variant M5 (WT + Q358N) showed 44.5-fold higher catalytic efficiency (k cat/K M) than BcLeuDH WT, which suggested that BcLeuDH M5 is an attractive candidate for L-ABA production on a large scale. Furthermore, the structure-functional relationship was investigated based on the docking and kinetic results.




Similar content being viewed by others
References
Zhu, L., Tao, R. S., Wang, Y., Jiang, Y., Lin, X., Yang, Y. L., Zheng, H. B., Jiang, W. H., & Yang, S. (2011). Removal of l-alanine from the production of l-2-aminobutyric acid by introduction of alanine racemase and D-amino acid oxidase. Applied Microbiology and Biotechnology, 90, 903–910.
Fotheringham, I. G., Grinter, N., Pantaleone, D. P., Senkpeil, R. F., & Taylor, P. P. (1999). Engineering of a novel biochemical pathway for the biosynthesis of L-2-aminobutyric acid in Escherichia coli K12. Bioorganic & Medicinal Chemistry, 7, 2209–2213.
Tao, R. S., Jiang, Y., Zhu, F. Y., & Yang, S. (2013). A one-pot system for production of L-2-aminobutyric acid from L-threonine by L-threonine deaminase and a NADH-regeneration system based on L-leucine dehydrogenase and formate dehydrogenase. Biotechnology Letters., 36, 1–7.
Xu, G., Jiang, Y., Tao, R. S., Wang, S. F., Zeng, H. Y., & Yang, S. (2016). A recyclable biotransformation system for l-2-aminobutyric acid production based on immobilized enzyme technology. Biotechnology Letters, 38, 123–129.
Zhu, W., Li, Y., Jia, H., Wei, P., Zhou, H., & Jiang, M. (2016). Expression, purification and characterization of a thermostable leucine dehydrogenase from the halophilic thermophile Laceyella sacchari. Biotechnology Letters, 38, 855–861.
Cheng, F., Zhu, L., & Schwaneberg, U. (2015). Directed evolution 2.0: improving and deciphering enzyme properties. Chemical Communications, 51, 9760–9672.
Jamil, S., Liu, M. H., Liu, Y. M., Han, R. Z., Xu, G. C., & Ni, Y. (2015). Hydrophobic mutagenesis and semi-rational engineering of arginine deiminase for markedly enhanced stability and catalytic efficiency. Applied Biochemistry & Biotechnology, 176, 1335–1350.
Xu, J. M., Fu, F. T., Hu, H. F., & Zheng, Y. G. (2016). A high-throughput screening method for amino acid dehydrogenase. Analytical Biochemistry, 495, 29–31.
Baker, P. J., Turnbull, A. P., Sedelnikova, S. E., Stillman, T. J., & Rice, D. W. (1995). A role for quaternary structure in the substrate-specificity of leucine dehydrogenase. Structure, 3, 693–705.
Zhao, Y., Wakamatsu, T., Doi, K., Sakuraba, H., & Ohshima, T. A. (2012). Psychrophilic leucine dehydrogenase from Sporosarcina psychrophila: purification, characterization, gene sequencing and crystal structure analysis. Journal of Molecular Catalysis B: Enzymatic, 83, 65–72.
Arnold, K., Bordoli, L., Kopp, J., & Schwede, T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics, 22, 195–201.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., & Higgins, D. G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948.
Cheng, F., Zhu, L., Lue, H., Bernhagen, J., & Schwaneberg, U. (2015). Directed arginine deiminase evolution for efficient inhibition of arginine-auxotrophic melanomas. Applied Microbiology and Biotechnology, 99, 1237–1247.
Liu, Z. Q., Baker, P. J., Cheng, F., Xue, Y. P., Zheng, Y. G., & Shen, Y. C. (2013). Screening and improving the recombinant nitrilases and application in biotransformation of iminodiacetonitrile to iminodiacetic acid. PloS One, 8, e67197.
Nagata, S., Bakthavatsalam, S., Galkin, A. G., Asada, H., Sakai, S., Esaki, N., Soda, K., Ohshima, T., Nagasaki, S., & Misono, H. (1995). Gene cloning, purification, and characterization of thermostable and halophilic leucine dehydrogenase from a halophilic thermophile, Bacillus licheniformis TSN9. Applied Microbiology & Biotechnology, 44, 432–438.
Li, H. M., Zhu, D. M., Hyatt, B. A., Malik, F. M., Biehl, E. R., & Hua, L. (2009). Cloning, protein sequence clarification, and substrate specificity of a leucine dehydrogenase from Bacillus sphaericus ATCC4525. Applied Biochemistry & Biotechnology, 158, 343–351.
Ohshima, T., Misono, H., & Soda, K. (1978). Properties of crystalline leucine dehydrogenase from Bacillus sphaericus. Journal of Biological Chemistry, 253, 5719–5725.
Ohshima, T., Nishida, N., Bakthavatsalam, S., Kataoka, K., Takada, H., Yoshimura, T., Esaki, N., & Soda, K. (1994). The purification, characterization, cloning and sequencing of the gene for a halostable and thermostable leucine dehydrogenase from Thermoactinomyces intermedius. European Journal of Biochemistry, 222, 305–312.
Shimoi, H., Nagata, S., Esaki, N., Tanaka, H., & Soda, K. (1987). Leucine dehydrogenase of a thermophilic anaerobe, Clostridium thermoaceticum: gene cloning, purification and characterization. Agricultural & Biological Chemistry, 51, 3375–3381.
Kärst, U., Schütte, H., Baydoun, H., & Tsai, H. (2004). Purification and characterization of leucine dehydrogenase from the thermophile ‘Bacillus caldolyticus’. Acta Polymerica Sinica, 37, 684–687.
Li, H., Zhu, D., Hyatt, B. A., Malik, F. M., Biehl, E. R., & Hua, L. (2009). Cloning, protein sequence clarification, and substrate specificity of a leucine dehydrogenase from Bacillus sphaericus ATCC4525. Applied Biochemistry & Biotechnology, 158, 343–351.
Schymkowitz, J., Borg, J., Stricher, F., Nys, R., Rousseau, F., & Serrano, L. (2005). The FoldX web server: an online force field. Nucleic Acids Research, 33, W382–W388.
Katoh, R., Nagata, S., & Misono, H. (2003). Cloning and sequencing of the leucine dehydrogenase gene from Bacillus sphaericus IFO 3525 and importance of the C-terminal region for the enzyme activity. Journal of Molecular Catalysis B: Enzymatic, 23, 239–247.
Zhu, L., Cheng, F., Piatkowski, V., & Schwaneberg, U. (2014). Protein engineering of the antitumor enzyme PpADI for improved thermal resistance. Chembiochem, 15, 276–283.
Acknowledgements
This work was financially supported by the National Major Scientific and Technological Special Project for “Significant Scientific Instrument and Equipment Development” (2012YQ15008713), the Zhejiang Provincial Natural Science Foundation of China (Q17C050006), and for Zhejiang Provincial Public Welfare Technology Application Research Projects (2017C33157).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Xu, JM., Cheng, F., Fu, FT. et al. Semi-Rational Engineering of Leucine Dehydrogenase for L-2-Aminobutyric Acid Production. Appl Biochem Biotechnol 182, 898–909 (2017). https://doi.org/10.1007/s12010-016-2369-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2369-0