Abstract
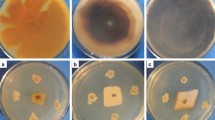
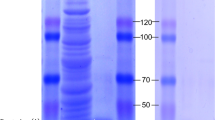
A novel β-1,3-glucanase gene, designated Ccglu17A, was cloned from the biological control fungus Chaetomium cupreum Ame. Its 1626-bp open reading frame encoded 541 amino acids. The corresponding amino acid sequence showed highest identity (67 %) with a glycoside hydrolase family 17 β-1,3-glucanase from Chaetomium globosum. The recombinant protein Ccglu17A was successfully expressed in Pichia pastoris, and the enzyme was purified to homogeneity with 10.1-fold purification and 47.8 % recovery yield. The protein’s molecular mass was approximately 65 kDa, and its maximum activity appeared at pH 5.0 and temperature 45 °C. Heavy metal ions Fe2+, Mn2+, Cu2+, Co2+, Ag+, and Hg2+ had inhibitory effects on Ccglu17A, but Ba2+ promoted the enzyme’s activity. Ccglu17A exhibited high substrate specificity, almost exclusively catalyzing β-1,3-glycosidic bond cleavage in various polysaccharoses to liberate glucose. The enzyme had a Km of 2.84 mg/mL and Vmax of 10.7 μmol glucose/min/mg protein for laminarin degradation under optimal conditions. Ccglu17A was an exoglucanase with transglycosylation activity based on its hydrolytic properties. It showed potential antifungal activity with a degradative effect on cell walls and inhibitory action against the germination of pathogenic fungus. In conclusion, Ccglu17A is the first functional exo-1,3-β-glucanase to be identified from C. cupreum and has potential applicability in industry and agriculture.





Similar content being viewed by others
References
Watanabe, T., Kasahara, N., Aida, K., & Tanaka, H. (1992). Three N-terminal domains of beta-1,3-glucanase A1 are involved in binding to insoluble beta-1,3-glucan. Journal of Bacteriology, 174(1), 186–190.
Ooi, T., Sato, H., Matsumoto, K., & Taguchi, S. (2009). A unique post-translational processing of an exo-beta-1,3-glucanase of Penicillium sp. KH10 expressed in Aspergillus oryzae. Protein Expression and Purification, 67(2), 126–131.
Genta, F. A., Bragatto, I., Terra, W. R., & Ferreira, C. (2009). Purification, characterization and sequencing of the major beta-1,3-glucanase from the midgut of Tenebrio molitor larvae. Insect Biochemistry and Molecular Biology, 39(12), 861–874.
Takeda, T., Nakano, Y., Takahashi, M., Konno, N., Sakamoto, Y., Arashida, R., et al. (2015). Identification and enzymatic characterization of an endo-1,3-beta-glucanase from Euglena gracilis. Phytochemistry, 116, 21–27.
Xie, Y. R., Raruang, Y., Chen, Z. Y., Brown, R. L., & Cleveland, T. E. (2015). ZmGns, a maize class I beta-1,3-glucanase, is induced by biotic stresses and possesses strong antimicrobial activity. Journal of Integrative Plant Biology, 57(3), 271–283.
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S. N., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Research, 43(D1), D222–D226.
Stahmann, K. P., Pielken, P., Schimz, K. L., & Sahm, H. (1992). Degradation of extracellular beta-(1,3)(1,6)-d-glucan by Botrytis cinerea. Applied and Environmental Microbiology, 58(10), 3347–3354.
Schaeffer, H. J., Leykam, J., & Walton, J. D. (1994). Cloning and targeted gene disruption of EXG1, encoding exo-beta 1, 3-glucanase, in the phytopathogenic fungus Cochliobolus carbonum. Applied and Environmental Microbiology, 60(2), 594–598.
Lin, C., Yang, J., Sun, H., Huang, X., Wang, R., & Zhang, K. Q. (2007). Purification and characterization of a beta-1,3-glucanase from the novel mycoparasite Periconia byssoides. Biotechnology Letters, 29(4), 617–622.
Wang, Y. J., & Yang, Q. (2009). Cloning and expression of a novel chitinase chi58 from Chaetomium cupreum in Pichia pastoris. Biochemical Genetics, 47(7–8), 547–558.
Zhang, H., & Li, M. (2012). Transcriptional profiling of ESTs from the biocontrol fungus Chaetomium cupreum. Scientific World Journal, 2012, 1–7.
Zhang, H., Yang, Q., Wang, G., & Shang, F. D. (2009). Analysis of expressed sequence tags from Chaetomium cupreum grown under conditions associated with mycoparasitism. Letters in Applied Microbiology, 48(3), 275–280.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10), 2731–2739.
Guo, W., Gonzalez-Candelas, L., & Kolattukudy, P. E. (1995). Cloning of a new pectate lyase gene pelC from Fusarium solani f. Sp. pisi (Nectria haematococca, mating type VI) and characterization of the gene product expressed in Pichia pastoris. Archives of Biochemistry and Biophysics, 323(2), 352–360.
Randtke, E. A., Chen, L. Q., Corrales, L. R., & Pagel, M. D. (2014). The Hanes-Woolf linear QUESP method improves the measurements of fast chemical exchange rates with CEST MRI. Magnetic Resonance in Medicine, 71(4), 1603–1612.
Sakamoto, Y., Nakade, K., & Konno, N. (2011). Endo-beta-1,3-glucanase GLU1, from the fruiting body of Lentinula edodes, belongs to a new glycoside hydrolase family. Applied and Environmental Microbiology, 77(23), 8350–8354.
Kawai, R., Igarashi, K., & Samejima, M. (2006). Gene cloning and heterologous expression of glycoside hydrolase family 55 beta-1,3-glucanase from the basidiomycete Phanerochaete chrysosporium. Biotechnology Letters, 28(6), 365–371.
Boonvitthya, N., Tanapong, P., Kanngan, P., Burapatana, V., & Chulalaksananukul, W. (2012). Cloning and expression of the Aspergillus oryzae glucan 1,3-beta-glucosidase a (exgA) in Pichia pastoris. Biotechnology Letters, 34(10), 1937–1943.
Guo, Y., Yan, Q. J., Yang, Y., Yang, S. Q., Liu, Y., & Jiang, Z. Q. (2015). Expression and characterization of a novel beta-glucosidase, with transglycosylation and exo-beta-1,3-glucanase activities, from Rhizomucor miehei. Food Chemistry, 175, 431–438.
Vijayendra, S. V. N., & Kashiwagi, Y. (2009). Characterization of a new acid stable exo-beta-1,3-glucanase of Rhizoctonia solani and its action on microbial polysaccharides. International Journal of Biological Macromolecules, 44(1), 92–97.
Takeuchi, K. (1983). Purification and characterization of exo-beta-1,3-glucanase from a hatching supernatant of Strongylocentrotus intermedius. Canadian Journal of Biochemistry and Cell Biology, 61(1), 54–62.
Bara, M. T., Lima, A. L., & Ulhoa, C. J. (2003). Purification and characterization of an exo-beta-1,3-glucanase produced by Trichoderma asperellum. FEMS Microbiology Letters, 219(1), 81–85.
Peng, Y., Liu, G. L., Yu, X. J., Wang, X. H., Jing, L., & Chi, Z. M. (2011). Cloning of Exo-beta-1,3-glucanase Gene from a marine yeast Williopsis saturnus and its overexpression in Yarrowia lipolytica. Marine Biotechnology, 13(2), 193–204.
Monteiro, V. N., & Ulhoa, C. J. (2006). Biochemical characterization of a beta-1,3-glucanase from Trichoderma koningii induced by cell wall of Rhizoctonia solani. Current Microbiology, 52(2), 92–96.
Sakdapetsiri, C., Fukuta, Y., Aramsirirujiwet, Y., Shirasaka, N., & Kitpreechavanich, V. (2016). Antagonistic activity of endo-beta-1,3-glucanase from a novel isolate, Streptomyces sp. 9X166, against black rot in orchids. Journal of Basic Microbiology, 56(5), 469–479.
Sun, H., Yang, J. K., Lin, C., Huang, X. W., Xing, R. H., & Zhang, K. Q. (2006). Purification and properties of a beta-1,3-glucanase from Chaetomium sp that is involved in mycoparasitism. Biotechnology Letters, 28(2), 131–135.
Liu, B., Lu, Y., Xin, Z., & Zhang, Z. (2009). Identification and antifungal assay of a wheat beta-1,3-glucanase. Biotechnology Letters, 31(7), 1005–1010.
Acknowledgements
The authors wish to thank the Chinese government for the financial support of this study under the National High Technology Research and Development Program (2011AA10A205) and “Twelfth Five-Year Plan” National Science and Technology Program on Rural Area (2014BAL02B00).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Jiang, C., Song, J., Cong, H. et al. Expression and Characterization of a Novel Antifungal Exo-β-1,3-glucanase from Chaetomium cupreum . Appl Biochem Biotechnol 182, 261–275 (2017). https://doi.org/10.1007/s12010-016-2325-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2325-z