Abstract
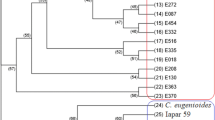
Understanding the genetic diversity of a crop is useful for its effective utilization in breeding programmes. For better understanding of the genetic variability in common bean, the first and foremost step is to study its genetic diversity. In the present investigation, 138 genotypes of common bean collected from various regions of Jammu and Kashmir, India, representing major common bean growing areas of this region, were evaluated using 23 SSRs. These SSRs were found highly polymorphic and possess high values for various parameters indicating their high discriminatory power. The average PIC value observed was 0.692, with 0.730 as average gene diversity value, and 0.267 as heterozygosity. Twenty-three SSRs produced a total of 251 alleles. The dendrogram generated with un-weighted neighbour joining cluster analysis grouped genotypes into three main clusters with various degrees of sub-clustering within the clusters. The model-based STRUCTURE analysis using 23 SSR markers identified a population with 3 sub-populations which corresponds to distance-based groupings with average F ST value and expected heterozygosity of 0.1497 and 0.6696, respectively, within the sub-population, as such high level of genetic diversity was observed within the population. Further, Core Hunter II was used to identify a core set of 96 diverse genotypes. This core set of diverse 96 genotypes is a potential resource for association mapping studies and can be used by breeders as a material to make desirable genetic crosses to generate elite varieties for the fulfilling global market needs. These findings have further implications in common bean breeding as well as conservation programs.


Similar content being viewed by others
References
Beebe, S., Gonzalez, A. V., & Rengifo, J. (2000). Research on trace minerals in the common bean. Food Nutrition Bulletin, 21, 387–391.
Svetleva, D., Pereira, G., Carlier, J., Cabrita, L., Leitao, J., & Genchev, D. (2006). Molecular characterization of Phaseolus vulgaris L. Genotypes included in Bulgarian collection by ISSR and AFLP analyses. Scientia Horticulturae, 109, 198–206.
Nyombaire, G., Siddiq, M., & Dolan, K. (2007). Effect of soaking and cooking on the oligosaccharides and lectins of red kidney beans (Phaseolus vulgaris L.). Bean Improv Coop Ann Rep, 50, 31–32.
Kaplan, L., & Lynch, T. F. Phaseolus (Fabaceae) in archaeology: AMS radiocarbon dates and their significance for pre-Colombian agriculture. Economic Botany, 53(3), 261–272.
Wortmann, C.S., Brink, M., Belay, G. (2006) Phaseolus vulgaris L. (Common bean). Record from PROTA4U. In PROTA (Plant Resources of Tropical Africa / Ressources végétales de l’Afrique tropicale) (Eds.), Wageningen, Netherlands.
McClean, P. E., Terpstra, J., McConnell, M., White, C., Lee, R., & Mamidi, S. (2012). Population structure and genetic differentiation among the USDA common bean (Phaseolus vulgaris L.) core collection. Genetic Resource and Crop Evolution, 59, 499–515.
Kaplan, L. (1981). What is the origin of the common bean? Economic Botany, 35, 240–254.
Singh, S. P., Nodari, R., & Gepts, P. (1991a). Genetic diversity in cultivated common bean. 1. Allozymes. Crop Science, 31, 19–23.
Galvan, M. Z., Mene’ndez-Sevillano, M. C., De Ron, A. M., Santalla, M., & Balatti, P. A. (2006). Genetic diversity among wild common beans from northwestern Argentina based on morpho agronomic and RAPD data. Genetic Resources and Crop Evolution, 53, 891–900.
Gepts, P., & Bliss, F. A. (1985). F1-hybrid weakness in the common bean—differential geographic origin suggests two gene pools in cultivated bean germplasm. Journal of Heredity, 76, 447–450.
Gepts, P., Osborn, T. C., Rashka, K., & Bliss, F. A. (1986). Phaseolin-protein variability in wild forms and landraces of the common bean (Phaseolus vulgaris). Evidence for multiple centers of domestication. Economic Botany, 40, 451–468.
Koenig, R., & Gepts, P. (1989). Allozyme diversity in wild Phaseolus vulgaris: further evidence for two major centers of genetic diversity. Theoretical and Applied Genetics, 78, 809–817.
Becerra, V., & Gepts, P. (1994). RFLP diversity of common bean (Phaseolus vulgaris L.) in its centres of origin. Genome, 37, 256–263.
Me’tais, I., Aubry, C., Hamon, B., Jalouzot, R., & Peltier, D. (2000). Description and analysis of genetic diversity between commercial bean lines (Phaseolus vulgaris L.). Theoretical and Applied Genetics, 101, 1207–1214.
Caicedo, A. L., Gaitan, E., Duque, M. C., Torochica, O., Debouck, D. G., & Thome, J. (1999). AFLP fingerprinting of Phaseolus lunatus L. And related wild species from S. America. Crop Science, 39, 1497–1507.
Fabio, L. M., Sergio, E., Lee, T. S. G., & Flipe, G. G. (2003). Genetic relationships and diversity among Brazilian cultivars and land races of common beans (Phaseolus vulgaris L.) revealed by AFLP markers. Genetic Resources and Crop Evolution, 50, 887–893.
Pallottini, L., Garcia, E., Kami, J., Barcaccia, G., & Gepts, P. (2004). The genetic anatomy of a patented yellow bean. Crop Science, 44, 968–977.
Rosales-Serna, R., Hernandez-Delgado, S., Gonzalez-Paz, M., Acosta-Gallegos, J. A., & Mayek-Perez, N. (2005). Genetic relationships and diversity revealed by AFLP markers in Mexican common bean bred cultivars. Crop Science, 45, 1951–1957.
Duarte, M. J., Bosco dos Santos, J., & Cunha Melo, L. (1999). Genetic divergence among common bean cultivars from different races based on RAPD markers. Genetics and Moecular Biology, 22, 419–426.
Beebe, S., Skroch, P. W., Tohme, J., Duque, M. C., Pedraza, F., & Nienhuis, J. (2000). Structure of genetic diversity among J., common bean landraces of middle American origin based on correspondence analysis of RAPD. Crop Science, 40, 264–273.
Maciel, F., Gerald, L. T. S., & Echeverrigaray, S. (2001). Random amplified polymorphic DNA (RAPD) markers variability among cultivars and landraces of common bean (P. vulgaris L.) of South Brazil. Euphytica, 120, 257–263.
Tiwari, M., Singh, N. K., Rathore, M., & Kumar, N. (2005). RAPD markers in the analysis of genetic diversity among common bean germplasm from central Himalaya. Genetic Resources and Crop Evolution, 52, 315–324.
Blair, M. W., Giraldo, M. C., Buendia, H. F., Tovar, E., Duque, M. C., & Beebe, S. E. (2006). Microsatellite marker diversity in common bean (Phaseolus vulgaris L.). Theoretical and Applied Genetics, 113, 100–109.
Zhang, X., Blair, M. W., & Wang, S. (2008). Genetic diversity of Chinese common bean (Phaseolus vulgaris L.) landraces assessed with simple sequence repeat (SSR) markers. Theoretical and Applied Genetics, 117, 629–640.
Blair, M. W., Diaz, L. M., Gill-Langarica, H. R., Rosales-Serna, R., Mayek-Perez, N., & Acosta-Gallegos, J. A. (2011a). Genetic relatedness of Mexican common bean cultivars revealed by microsatellite markers. Crop Science, 51, 2655–2667.
Blair, M. W., Cortes, A. J., & Chavarro, M. C. (2011b). SNP marker diversity in common bean (Phaseolus vulgaris L.). Theoretical and Applied Genetics, 123, 827–845.
Dı’az, L. M., & Blair, M. W. (2006). Race structure within the Mesoamerican gene pool of common bean (Phaseolus vulgaris L.) as determined by microsatellite markers. Theoretical and Applied Genetics, 114, 143–154.
Asfaw, A., Blair, M. W., & Almekinders, C. (2009). Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) landraces from the east African highlands. Theoretical and Applied Genetics, 120, 1–12.
Blair, M. W., Gonzales, L. F., Kimani, P., & Butare, L. (2010). Inter-genepool introgression, genetic diversity and nutritional quality of common bean (Phaseolus vulgaris L.) landraces from Central Africa. Theoretical and Applied Genetics, 121, 237–248.
Burle, M. L., Fonseca, J. R., Kami, J. A., & Gepts, P. (2010). Microsatellite diversity and genetic structure among common bean (Phaseolus vulgaris L.) landraces in Brazil, a secondary center of diversity. Theoretical and Applied Genetics, 121, 801–813.
Xu, S., Wang, G., Mao, W., Hu, Q., Liu, N., Ye, L., & Gong, Y. (2014). Genetic diversity and population structure of common bean (Phaseolus vulgaris) landraces from China revealed by a new set of EST-SSR markers. Biochemical Systematics and Ecology, 57, 250–256.
Masi, P., Spagnoletti, Z. P., & Donini, P. (2003). Development and analysis of multiplex microsatellite markers sets in common bean (Phaseolus vulgaris L.). Molecular Breeding, 11, 303–313.
Me’tais, I., Hamon, B., Jalouzot, R., & Peltier, D. (2002). Structure and level of genetic diversity in various bean types evidenced with microsatellite markers isolated from a genomic enriched library. Theoretical and Applied Genetics, 104, 1346–1352.
Gome’z, O., Blair, M. W., Frankow-Lindberg, B., & Gullberg, U. (2004). Molecular and phenotypic diversity of common bean landraces from Nicaragua. Crop Science, 4, 1412–1418.
Dı’az, L. M., Buendı’a, H. F., Duque, M. C., & Blair, M. W. (2011). Genetic diversity of Colombian landraces of common bean as detected through the use of silver-stained and fluorescently labeled microsatellites. Plant Genetic Resources, 9, 86–96.
Payro dela Cruz, P., Gepts, P., Garcia Marı’n, P. C., & Villareal, D. Z. (2005). Spatial distribution of genetic diversity in wild populations of Phaseolus vulgaris L. From Guanajuato and Michoaca’n, Me’xico. Genetic Resources and Crop Evolution, 52, 589–599.
Hegay, S., Geleta, M., Bryngelsson, T., Gustavsson, L., Hovmalm, H. P., & Ortiz, R. (2012). ISSN 2322-1690 Comparing genetic diversity and population structure of common beans grown in Kyrgyzstan using microsatellites. Scientific Journal of Crop Science, 1(4), 63–75.
Scaranoa, D., Fernando, R., José, R. J., Rosa, R., & Giandomenico, C. (2014). Morphological and genetic diversity among and within common bean (Phaseolus vulgaris L.) landraces from the Campania region (southern Italy). Scientia Horticulturae, 180, 72–78.
Kumar, V., Sharma, S., Sharma, A.K., Sharma, S., Bhat, K.V. (2009). Comparative analysis of diversity based on morpho-agronomic traits and microsatellite markers in common bean. Euphytica, 170 (3): 249–262.
Sharma, P. N., Dı’az, L. M., & Blair, M. W. (2013). Genetic diversity of two Indian common bean germplasm collections based on morphological and microsatellite markers. Plant Genetic Resource, 11(2), 121–130.
Gopinath, S. M., Katti, A. V., Dayananda, K. S., Shareef, M. I., Nair, D. V. (2013). Assessment of genetic diversity of French bean using SSR primers. International Journal of Innovative Research in Science, Engineering and Technology, 2 (9), 4745–4752.
Zargar, S. M., Farhat, S., Mahajan, R., Bhakhri, A., Sharma, A. (2016). Unraveling the efficiency of RAPD and SSR markers in diversity analysis and population structure estimation in common bean. Saudi Journal of Biological Sciences, 23 (1), 139–149.
Pritchard, J. K., Stephens, M., & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959.
Thornsberry, J. M., Goodman, M. M., Doebley, J., Kresovich, S., Nielsen, D., & Buckler, E. S. (2001). Dwarf 8 polymorphisms associate with variation in flowering time. Nature Genetics, 28, 286–289.
Pritchard, J. K., Stephens, M., Rosenberg, N. A., & Donnelly, P. (2000b). Association mapping in structured populations. The American Journal of Human Genetics, 67, 170–181.
Pritchard, J. K., & Rosenberg, N. A. (1999). Use of unlinked genetic markers to detect population stratification in association studies. The American Journal of Human Genetics, 65, 220–228.
Doyle, J. J., & Doyle, J. L. (1987). A rapid DNA isolation procedure to small amounts of fresh leaf tissue. Phytochemistry Bulletin, 19, 11–15.
Yu, K., Park, S. J., Poysa, V., & Gepts, P. (2000). Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.). Heredity, 91, 429–434.
Gaitan-Solis, E., Duque, M. C., Edwards, K. J., & Tohme, J. (2002). Microsatellite repeats in common bean (Phaseolus vulgaris): isolation, characterization, and cross-species amplification in Phaseolus ssp. Crop Science, 42, 2128–2136.
Grisi, M. C. M., Blair, M. W., Gepts, P., Brondani, C., Pereira, P. A. A., & Brondadi, R. P. V. (2007). Genetic mapping of a new set of microsatellite markers in a reference common bean (Phaseolus vulgaris) population BAT93 x Jalo EEP558. Genetics and Molecular Research, 3, 691–706.
Hanai, L. R., Santini, L., Camargo, L. E. A., Fungaro, M. H. P., Gepts, P., Tsai, S. M., & Vieira, M. L. C. (2010). Extension of the core map of common bean with EST-SSR, RGA, AFLP and putative functional markers. Molecular Breeding, 25(1), 25–45.
Co’ rdoba, J. M., Chavarro, C., Schlueter, J. A., Jackson, S. A., & Blair, M. W. (2010). Integration of physical and genetic maps of common bean through BAC-derived microsatellite markers. BMC Genomics, 11, 436.
Bassam, B. J., Caetano-Anolles, G., & Gresshoffer, P. M. (1991). Fast and sensitive silver staining of DNA in polyacrylamide gels. Annals of Biochemistry, 196(1), 80–83.
Botstein, D., White, R. L., Skolnick, M., & Davis, R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics, 32, 314–331.
Nei, M. (1987). Molecular evolutionary genetics. New York: Columbia University press.
Liu, K., & Muse, S. V. (2005). Power marker: an integrated analysis environment for genetic marker analysis. Bioinformatics, 21(9), 2128–2129.
Perrier, X., & Jacquemoud-Collet, J.P. (2006). DARwin software. http://darwin.cirad.fr/darwin.
Nei, M. (1972). Genetic distance between populations. American Naturalist, 106, 283–292.
Evano, G., Regnaut, S., & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14, 2611–2620.
Earl, D., & von Holdt, B. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetic Resources, 4(2), 359–361.
Jia, Y., Sun, J., Wang, X., Zhou, Z., Pan, Z., He, S., et al. (2013). Molecular diversity and association analysis of drought and salt tolerance in G. hirsutum L. Germplasm. Journal of Integrative Agriculture Advanced. doi:10.1016/S2095-3119(13)60668-1.
Jakobsson, M., & Rosenberg, N. A. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23(14), 1801–1806.
Ramasamy, R. K., Ramasamy, S., Bindroo, B. B., & Naik, V. G. (2014). STRUCTURE PLOT: a program for drawing elegant STRUCTURE bar plots in user friendly interface. Springerplus, 13(3), 431.
Wright, S. (1951). The genetical structure of populations. Annals of Eugenics, 15, 323–354.
De Beukelaer, H., Sm’ykal, P., Davenport, G. F., & Fack, V. (2012). Core hunter II: fast core subset selection based on multiple genetic diversity measures using mixed replica search. BMC Bioinformatics, 13, 312.
lair, M. W. B., Soler, A., & Corte’s, A. J. (2012). Diversification and population structure in common beans (Phaseolus vulgaris L.). PloS One, 7(11), e49488.
Jime’nez, O. R., & Korpelainen, H. (2012). Microsatellite markers reveal promising genetic diversity and seed trait associations in common bean landraces (Phaseolus vulgaris L.) from Nicaragua. Plant Genetic Resources, 10(2), 108–118.
Maras, M., Susˇtar-Vozlicˇ, J., Javornik, B., & Meglicˇ, V. (2008). The efficiency of AFLP and SSR markers in genetic diversity estimation and gene pool classification of common bean (Phaseolus vulgaris L.). Acta Agriculturae Slovenica, 91, 87–96.
Okii, D., Tukamuhabwa, P., Kami, J., Namayanja, A., Paparu, P., Ugen, M., & Gepts, P. (2014). The genetic diversity and population structure of common bean (Phaseolus vulgaris L) germplasm in Uganda. African Journal of Biotechnology, 13(29), 2935–2949.
Singh, S. P., Gutie’rrez, J. A., Molina, A., Urrea, C., & Gepts, P. (1991b). Genetic diversity in cultivated common bean. II. Marker-based analysis of morphological and agronomic traits. Crop Science, 31, 23–29.
Logozzo, G., Donnoli, R., Macaluso, L., Papa, R., Knüpffer, H., & Zeuli, P. S. (2007). Analysis of the contribution of Mesoamerican and Andean gene pools to European common bean (Phaseolus vulgaris L.) germplasm and strategies to establish a core collection. Genetic Resources and Crop Evolution, 54, 1763–1779.
Perseguini, J. M. K. C., Silva, G. M. B., Rosa, J. R. B. F., Gazaffi, R., Marçal, J. F., Carbonell, S. A. M., et al. (2015). Developing a common bean core collection suitable for association mapping studies. Genetics and Molecular Biology, 38(1), 67–78.
Warburton, M., Crossa, J., Diaz, L., Gomez, A., Taba, S. (2004). Diversidad gen’ etica en criollos demais medida pormicrosat ‘ elites. In Congreso Nacional de Biotecnolog’ıa Agropecuaria y Forestal. Chapingo, M’exico.
Dubreuil, P., Warburton, M., Chastanet, M., Hoisington, D., & Charcosset, A. (2006). More on the introduction of temperate maize into Europe: large-scale bulk SSR genotyping and new historical elements. Maydica, 51, 281–291.
Sm’ykal, P., Baˇcov’a-Kerteszov’a, N., Kalendar, R., Corander, J., Schulman, A. H., & Pavelek, M. (2011a). Genetic diversity of cultivated flax (Linum usitatissimum L.) germplasm assessed by retrotransposon-basedmarkers. Theoretical and Applied Genetics, 122, 1385–1397.
Sm’ykal, P., H’ybl, M., Corander, J., Jarkovsk’y, J., Flavell, A., & Griga, M. (2008). Genetic diversity and population structure of pea (Pisum sativum L.) varieties derived from combined retrotransposon, microsatellite and morphological marker analysis. Theoretical and Applied Genetics, 117, 413–424.
Jing, R., Vershinin, A., Grzebyta, J., Shaw, P., Sm’ykal, P., Marshall, D., et al. (2010). The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis. BMC Evolutionary Biology, 10, 44.
Sm’ykal, P., Kenicer, G., Flavell, A. J., Corander, J., Kosterin, O., Redden, R. J., et al. (2011b). Phylogeny, phylogeography and genetic diversity of the Pisum genus. Plant Genetic Resource, 9, 4–18.
Acknowledgements
SMZ is grateful to SERB, DST, New Delhi for financial support of this work (Project sanction order No. SR/FT/LS-27/ 2011).
Contribution of Authors
RM was involved in laboratory experiments and was also involved in data analysis. SMZ has designed experiment and has guided molecular experiments, analysis and manuscript preparation. RS has helped in data analysis and manuscript correction. SF and HS helped in data analysis. RKS helped in manuscript preparation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors have no conflict of interest.
Submission Declaration
The work described has neither been published nor is under consideration for publication in any other journal, and if accepted, it will not be published anywhere in this form or any other form.
Electronic supplementary material
Supplementary Figure1
Jammu and Kashmir Map. Red triangle indicates the region of common bean collection. (PPTX 155 kb)
Supplementary Figure 2
NJ tree dendrogram. Red colour indicates population I, green colour indicates population II and Blue colour indicates population III and grey colour admixture. (PPTX 84 kb)
Supplementary Figure 3
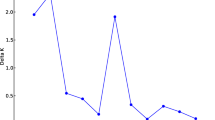
Graphical representation of the optimal number of groups in STRUCTURE inferred using the criterion of Evanno et al. (2005). The analysis was based on data obtained using 23 microsatellite markers. (PPTX 122 kb)
Supplementary Figure 4
Core Set of 96 diverse common bean genotypes. (PPTX 114 kb)
Supplementary Table 1
(DOCX 21 kb)
Supplementary Table 2
(DOCX 25 kb)
Rights and permissions
About this article
Cite this article
Mahajan, R., Zargar, S.M., Singh, R. et al. Population Structure Analysis and Selection of Core Set among Common Bean Genotypes from Jammu and Kashmir, India. Appl Biochem Biotechnol 182, 16–28 (2017). https://doi.org/10.1007/s12010-016-2307-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2307-1