Abstract
Determination of genetic stability of in vitro-grown plantlets is needed for safe and large-scale production of mature trees. In this study, genetic variation of long-term micropropagated mature pistachio developed through direct shoot bud regeneration using apical buds (protocol A) and in vitro-derived leaves (protocol B) was assessed via DNA-based molecular markers. Randomly amplified polymorphic DNA (RAPD), inter-simple sequence repeat (ISSR), and amplified fragment length polymorphism (AFLP) were employed, and the obtained PIC values from RAPD (0.226), ISSR (0.220), and AFLP (0.241) showed that micropropagation of pistachio for different periods of time resulted in “reasonable polymorphism” among donor plant and its 18 clones. Mantel’s test showed a consistence polymorphism level between marker systems based on similarity matrices. In conclusion, this is the first study on occurrence of genetic variability in long-term micropropagated mature pistachio plantlets. The obtained results clearly indicated that different marker approaches used in this study are reliable for assessing tissue culture-induced variations in long-term cultured pistachio plantlets.


Similar content being viewed by others
Abbreviations
- AFLP:
-
Amplified fragment length polymorphism
- BA:
-
6-Benzyladenine
- IAA:
-
Indole acetic acid
- IBA:
-
Indole-3-butyric acid
- EMR:
-
Effective multiplex ratio
- Ib av :
-
Average band informativeness
- ISSR:
-
Inter-simple sequence repeat
- MI:
-
Marker index
- PCA:
-
Principal component analysis
- PC:
-
Principal coordinate
- PIC:
-
Polymorphic information content
- RAPD:
-
Randomly amplified polymorphic DNA
- RFLP:
-
Restriction fragment length polymorphism
- R p :
-
Resolving power
- SSR:
-
Simple sequence repeat
- TC:
-
Tissue culture
- UPGMA:
-
Unweighted pair-group method with arithmetic
References
Ozden-Tokatli, Y., Akdemir, H., Tilkat, E., & Onay, A. (2010). Current status and conservation of Pistacia germplasm. Biotechnology Advances, 28, 130–141.
Tilkat, E., & Onay, A. (2009). Direct shoot organogenesis from in vitro derived mature leaf explants of pistachio. In Vitro Cellular & Developmental Biology. Plant, 45(1), 92–98.
Yadav, K., Aggarwal, A., & Singh, N. (2013). Evaluation of genetic fidelity among micropropagated plants of Gloriosa superba L. using DNA-based markers—a potential medicinal plant. Fitoterapia, 89, 265–270.
Barghchi, M., & Alderson, P. G. (1985). In vitro propagation of P. vera L. and commercial varieties of Ohadi and Kalleghochi. The Journal of Horticultural Science and Biotechnology, 60, 423–440.
Parfitt, D. E., & Almehdi, A. (1994). Use of high CO2 atmospheric and medium modifications for the successful micropropagation of pistachio. Scientia Horticulturae, 56, 321–329.
Onay, A. (2000). Micropropagation of pistachio from mature trees. Plant Cell, Tissue and Organ Culture, 60, 159–162.
Tilkat, E. (2006). Micropropagation of male Pistacia vera L. via apical shoot tip culture. Ph.D. Thesis Institute of Science, University of Dicle, Turkey, 142 p. (in Turkish).
Tilkat, E., Onay, A., Yıldırım, H., & Ozen, H. C. (2008). Micropropagation of mature male pistachio Pistacia vera L. The Journal of Horticultural Science and Biotechnology, 83(3), 328–333.
Tilkat, E., Onay, A., Yıldırım, H., & Ayaz, E. (2009). Direct plant regeneration from mature leaf explants of pistachio, Pistacia vera L. Scientia Horticulturae, 121(3), 361–365.
Orbovic, V., Calovic, M., Viloria, Z., Nielsen, B., Gmitter, F., Castle, W., & Grosser, J. (2008). Analysis of genetic variability in various tissue culture-derived lemon plant populations using RAPD and flow cytometry. Euphytica, 161, 329–335.
Leva, A.R., Petruccelli, R. &, Rinaldi, L. M. R. (2012). Somaclonal variation in tissue culture: a case study with olive. Recent advances in plant in vitro culture, 10–17.
Bhatia, R., Singh, K. P., Jhang, T., & Sharma, T. R. (2009). Assessment of clonal fidelity of micropropagated gerbera plants by ISSR markers. Scientia Horticulturae, 119, 208–211.
Sharma, M. M., Verma, R. N., Singh, A., & Batra, A. (2014). Assessment of clonal fidelity of Tylophora indica (Burm. f.) Merrill “in vitro” plantlets by ISSR molecular markers. Springer Plus, 3, 400.
Rathore, M. S., Yadav, P., Mastan, S. G., Prakash, C. R., Singh, A., & Agarwal, P. K. (2014). Evaluation of genetic homogeneity in tissue culture regenerates of Jatropha curcas L. using flow cytometer and DNA-based molecular markers. Applied Biochemistry and Biotechnology, 172, 298–310.
Bairu, M. W., Aremu, A. O., & Van Standen, J. (2011). Somaclonal variation in plants: causes and detection methods. Plant Growth Regulation, 63, 147–173.
Polanco, C., & Ruiz, M. L. (2002). AFLP analysis of somaclonal variation in Arabidopsis thaliana regenerated plants. Plant Science, 162, 817–824.
Prado, M., Rodriguez, E., Rey, L., Gonzalez, M., Santos, C., & Rey, M. (2010). Detection of somaclonal variants in somatic embryogenesis regenerated plants of Vitis vinifera by flow cytometry and microsatellite markers. Plant Cell, Tissue and Organ Culture, 103, 49–59.
Reddy, M. P., Sarla, N., & Siddiq, E. A. (2002). Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica, 128, 9–17.
Viehmannova, I., Bortlova, Z., Vitamvas, J., Cepkova, P. H., Eliasova, K., Svobodava, E., & Travnickova, M. (2014). Assessment of somaclonal variation in somatic embryo-derived plants of yacon [Smallanthus sonchifolius (Poepp. and Endl.) H. Robinson] using inter simple sequence repeat analysis and flow cytometry. Electronic Journal of Biotechnology, 17, 102–106.
Henry, R. J. (1998). Molecular and biochemical characterization of somaclonal variation. In S. M. Jain, D. S. Brar, & B. S. Ahloowalia (Eds.), Somaclonal variation and induced mutations in crop improvement (pp. 485–499). Dordrecht: Kluwer Acad. Publ.
Ozden-Tokatli, Y., Ozudogru, E. A., & Akcin, A. (2006). Optimization of an efficient micropropagation protocol and assessment of plant genetic fidelity by RAPD markers in pistachio (Pistacia vera). Scientia Horticulturae, 20(2), 162–169.
Murashige, T., & Skoog, M. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiologia Plantarum, 15, 473–497.
Lodhi, M. A., Guang-Ning, Y., Weeden, N. F. N., & Reisch, B. I. (1994). A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Molecular Biology Reports, 6–13.
Kovach, W. L. (1999). MVSP—a multivariate statistical package for Windows, ver 3.1. Pentraeth: Kovach Computing Services.
Jaccard, P. (1908). Nouvelles recherches sur la distribution florale. Bulletin de la Societe Vaudoise des Sciences Naturelles, 44, 223–270.
Rohlf, F. J. (1993). NTSYS-pc: numerical taxonomy and multivariate analysis system. New York: Exeter Publishing Ltd.
Roldan-Ruiz, I., Calsyn, E., Gilliland, T. J., Coll, R., van Eijk, M. J. T., & De Loose, M. (2000). Estimating genetic conformity between related ryegrass (Lolium) varieties. 2. AFLP characterization. Molecular Breeding, 6, 593–602.
Prevost, A., & Wilkinson, M. J. (1999). A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theoretical and Applied Genetics, 98, 107–112.
Belaj, A., Satovic, Z., Cipriani, G., Baldoni, L., Testolin, R., Rallo, L., & Trujillo, I. (2003). Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theoretical and Applied Genetics, 107(4), 736–744.
Campbell, B. C., LeMare, S., Piperidis, G., & Godwin, I. D. (2010). IRAP, a retrotransposon-based marker system for the detection of somaclonal variation in barley. Molecular Breeding, 27(2), 193–206.
Legendre, P., & Legendre, L. (1998). Numerical ecology, 24.
Jain, S. M. (1997). Micropropagation of selected somaclones of Begonia and Saintpaulia. Journal of Biosciences, 22(5), 585–592.
Devarumath, R. M., Nandy, S., Rani, V., Marimuthu, S., & Muraleedharan, N. (2002). RAPD, ISSR and RFLP fingerprints as useful markers to evaluate genetic integrity of micropropagated plants of three diploid and triploid elite tea clones representing Camellia sinensis (China type) and C. assamica ssp. assamica (Assam-India type). Plant Cell Reports, 21(2), 166–173.
Mallon, R., Rodríguez-Oubiña, J., & González, M. L. (2010). In vitro propagation of the endangered plant Centaurea ultreiae: assessment of genetic stability by cytological studies, flow cytometry and RAPD analysis. Plant Cell, Tissue and Organ Culture, 101, 31–39.
Rival, A., Bertrand, L., Beule, T., Combes, M. C., Trouslot, P., & Lashermes, P. (1998). Suitability of RAPD analysis for the detection of somaclonal variants in oil palm (Elaeis guineensis Jacq). Plant Breeding, 117, 73–76.
Sahijram, L., Soneji, J. R., & Bollamma, K. T. (2003). Analyzing somaclonal variation in micropropagated bananas (Musa spp.). In Vitro Cellular & Developmental Biology. Plant, 39, 551–556.
Martins, M., Sarmento, D., & Oleveira, M. M. (2004). Genetic stability of micropropagted almond plantlets as assessed by RAPD and ISSR markers. Plant Cell Reports, 23, 492–496.
Venkatachalam, L., Sreedhar, R. V., & Bhagyalakshmi, N. (2007). Molecular analysis of genetic stability in long-term micropropagated shoots of banana using RAPD and ISSR markers. Electronic Journal of Biotechnology, 15, 106–113.
Huang, W. J., Ning, G. G., Liu, G. F., & Bao, M. Z. (2009). Determination of genetic stability of long-term micropropagated plantlets of Platanus acerifolia using ISSR markers. Biologia Plantarum, 53(1), 159–163.
Rani, V., Parida, A., & Raina, S. N. (1995). Random amplified polymorphic DNA (RAPD) markers for genetic analysis in micropropagated plants of Populus deltoides Marsh. Plant Cell Reports, 14, 459–462.
Rahman, M., & Rajora, O. (2001). Microsatellite DNA somaclonal variation in micropropagated trembling aspen (Populus tremuloides). Plant Cell Reports, 20(6), 531–536.
Rani, V., Singh, K. P., Shiran, B., Goel, S. N. S., Devarumath, R. M., Sreenath, H. L., & Raina, S. N. (2000). Evidence for new nuclear and mitochondrial genome organizations among high-frequency somatic embryogenesis-derived plants of allotetraploid Coffea arabica L. (Rubiaceae). Plant Cell Reports, 19, 1013–1020.
Rani, V., & Raina, S. N. (1998). Genetic analysis of enhanced-axillary-branching-derived Eucalyptus tereticornis Smith and E. camaldulensis Dehn. plants. Plant Cell Reports, 17, 236–242.
Ngezahayo, F., Guo, W., Gong, L., Li, F., & Liu, B. (2006). Genomic variation in micropropagated Robinia ambigua ‘idahoensis’ revealed by RAPD markers. HortScience, 41(6), 1466–1468.
Guo, W. L., Gong, L., Ding, Z. F., Li, Y. D., Li, F. X., Zhao, S. P., & Liu, B. (2006). Genomic instability in phenotypically normal regenerants of medicinal plant Codonopsis lanceolata Benth. et Hook. F., as revealed by ISSR and RAPD markers. Plant Cell Reports, 25, 896–906.
Olmos, S. E., Lavia, G., Di Renzo, M., Mroginski, L., & Echenique, V. (2002). Genetic analysis of variation in micropropagated plants of Melia azedarach L. In Vitro Cellular and Development Biology - Plant, 38(6), 617–622.
Rajpal, V. R., Sharma, S., Devarumath, R. M., Chaudhary, M., Kumar, A., Khare, N., & Raina, S. N. (2014). In K. G. Ramawat, J. M. Mérillon, & M. R. Ahuja (Eds.), Tree biotechnology (pp. 303–334). FL: CRC Press.
Palombi, M. A., & Damiano, C. (2002). Comparison between RAPD and SSR molecular markers in detecting genetic variation in kiwifruit (Actinidia deliciosa A. Chev). Plant Cell Reports, 20(11), 1061–1066.
Agarwal, M., Shrivastava, N., & Padh, H. (2008). Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Reports, 27, 617–631.
Botstein, D., White, R. L., Skolnick, M., & Davis, R. W. (1980). Construction of a genetic linkage map using restriction fragment length polymorphisms. American Journal of Human Genetics, 32, 314–331.
Duncan, R. R. (1997). Tissue culture-induced variation and crop improvement. Advances in Agronomy, 58, 201–240.
Sharma, S., Bryan, G., Winfield, M., & Millam, S. (2007). Stability of potato (Solanum tuberosum L.) plants regenerated via somatic embryos, axillary bud proliferated shoots, microtubers and true potato seeds: a comparative phenotypic, cytogenetic and molecular assessment. Planta, 226, 1449–1458.
Acknowledgments
The authors are grateful to MSc. Emrah Kirdok for helping to statistical analysis. This research was funded by a grant no. KBAG 209 T030 from The Scientific and Technological Research Council of Turkey (TUBITAK).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supply. Fig. 1
Diagram of plant regeneration protocol A and B for mature P. vera L. (JPG 105 kb)
Supply. Fig. 2
All PCR programmes used in this study. (DOCX 218 kb)
Supply. Fig. 3
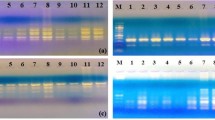
Examples of RAPD patterns assessed with K-01 primer; lanes D: Donor plant, G: Greenhouse material, 1-10: 5-years micropropagated plants, 20-26: 7-years micropropagated plants, “M: marker (some polymorphic bands were shown with arrowheads)”. (DOCX 281 kb)
Supply. Fig. 4
Examples of ISSR patterns assessed with ISSR-12 primer; lanes D: Donor plant, G: Greenhouse material, 1-10: 5-year micropropagated plants, 20-26: 7-years micropropagated plants, “M: marker (some polymorphic bands were shown with arrowheads)”. (DOCX 374 kb)
Supply. Fig. 5
Examples of AFLP patterns assessed with E-AAC and M-CTA primer combinations; lanes D: Donor plant, G: Greenhouse material, 1-10: 5-year micropropagated plants, 20-26: 7-years micropropagated plants, “M: marker (some polymorphic bands were shown with arrowheads)”. (DOCX 460 kb)
Rights and permissions
About this article
Cite this article
Akdemir, H., Suzerer, V., Tilkat, E. et al. Detection of Variation in Long-Term Micropropagated Mature Pistachio via DNA-Based Molecular Markers. Appl Biochem Biotechnol 180, 1301–1312 (2016). https://doi.org/10.1007/s12010-016-2168-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2168-7