Abstract
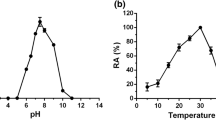
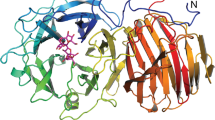
β-Fructosidase, a glycoside hydrolase of a biotechnologically important strain, was studied for its biochemical, physicochemical, and three-dimensional structure characteristics. This enzyme was heterologously expressed in Escherichia coli as a C-terminal His-tagged protein (SacB). β-Fructosidase catalyzes the cleavage of glycoside bonds toward certain carbohydrates with β-fructofuranosyl linkages; however, SacB exhibited selectivity toward sucrose and an optimum activity at pH 6.0–6.5 and 37 °C. In such optimum enzymatic activity conditions, the SacB was commonly observed as a monodisperse protein by dynamic light scattering (DLS). As β-fructosidase belongs to glycoside hydrolase family 32 (GH32), a β-sandwich and a five-bladed β-propeller domain are typical predicted folds in its structure. Docking and molecular dynamic simulations revealed for the first time a funnel-like channel perfectly exposed in the β-propeller domain of the Lactobacillus plantarum β-fructosidase (this allows the interaction between its entire catalytic triad and substrates that are larger than sucrose). In contrast, SacB showed a closed central tunnel collaterally induced by its His-tag.








Similar content being viewed by others
References
Lombard, V., Ramulu, H. G., Drula, E., Coutinho, P. M., & Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Research, 42(D1), D490–D495.
Nadeem, H., Rashid, M. H., Siddique, M. H., Azeem, F., Muzammil, S., Javed, M. R., Ali, M. A., Rasul, I., & Riaz, M. (2015). Microbial invertases: a review on kinetics, thermodynamics, physiochemical properties. Process Biochemistry, 50(8), 1202–1210.
Awad, G. E. A., Amer, H., El-Gammal, E. W., Helmy, W. A., & Elnashar, M. M. M. (2013). Production optimization of invertase by Lactobacillus brevis Mm-6 and its immobilization on alginate beads. Carbohydrate Polymers, 93(2), 740–746.
Nadeem, H., Rashid, M. H., Riaz, M., Asma, B., Javed, M. R., & Perveen, R. (2009). Invertase from hyper producer strain of Aspergillus niger: physiochemical properties, thermodynamics and active site residues heat of ionization. Protein and Peptide Letters, 16(9), 1098–1105.
Andjelković, U., Milutinović-Nikolić, A., Jović-Jovičić, N., Banković, P., Bajt, T., Mojović, Z., Vujčić, Z., & Jovanović, D. (2015). Efficient stabilization of Saccharomyces cerevisiae external invertase by immobilisation on modified beidellite nanoclays. Food Chemistry, 168(1), 262–269.
Diario Oficial de la Federación, Secretaría de Gobernación, México. Available from www.dof.gob.mx/nota_detalle.php?codigo=5413779&fecha=30/10/2015. Accessed 9 Feb 2016.
Jedrzejczak-Krzepkowska, M., Tkaczuk, K. L., & Bielecki, S. (2011). Biosynthesis, purification and characterization of β-fructofuranosidase from Bifidobacterium longum KN29.1. Process Biochemistry, 46(10), 1963–1972.
Ehrmann, M. A., Korakli, M., & Vogel, R. F. (2003). Identification of the gene for β-fructofuranosidase of Bifidobacterium lactis DSM10140T and characterization of the enzyme expressed in Escherichia coli. Current Microbiology, 46(6), 391–397.
Martel, C. M., Warrilow, A. G. S., Jackson, C. J., Mullins, J. G. L., Togawa, R. C., Parker, J. E., Morris, M. S., Donnison, I. S., Kelly, D. E., & Kelly, S. L. (2010). Expression, purification and use of the soluble domain of Lactobacillus paracasei β-fructosidase to optimise production of bioethanol from grass fructans. Bioresource Technology, 101(12), 4395–4402.
Chen, C., Zhou, F., Ren, J., Ai, L., Dong, Y., Wu, Z., Liu, Z., Chen, W., & Guo, B. (2014). Cloning, expression and functional validation of a β-fructofuranosidase from Lactobacillus plantarum. Process Biochemistry, 49(5), 758–767.
Kleerebezem, M., Boekhorst, J., van Kranenburg, R., Molenaar, D., Kuipers, O. P., Leer, R., Tarchini, R., Peters, S. A., Sandbrink, H. M., Fiers, M. W. E. J., Stiekema, W., Lankhorts, R. M. K., Bron, P. A., Hoffer, S. M., Groot, M. N. N., Kerkhoven, R., de Vries, M., Ursing, B., de Vos, W. M., & Siezen, R. J. (2003). Complete genome sequence of Lactobacillus plantarum WCFS1. Proceedings of the National Academy of Sciences USA, 100(4), 1990–1995.
Arena, M. P., Fiocco, D., Massa, S., Capozzi, V., Russo, P., & Spano, G. (2014). Lactobacillus plantarum as a strategy for an in situ production of vitamin B2. Journal of Food and Nutritional Disorders, S1-004.
Siezen, R. J., & van Hylckama Vlieg, J. E. (2011). Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microbial Cell Factories, 10(Suppl 1), S3.
Lamontanara, A., Caggianiello, G., Orrù, L., Capozzi, V., Michelotti, V., Bayjanov, J. R., Renckens, B., van Hijum, S. A. F. T., Cattivelli, L., & Spano, G. (2015). Draft genome sequence of Lactobacillus plantarum Lp90 isolated from wine. Genome Announcements, 3(2), e00097–15.
Sorger, P. K., & Pelham, H. R. B. (1987). Purification and characterization of a heat-shock element binding protein from yeast. The EMBO Journal, 6(10), 3035–3041.
Gao, J., Xu, Y.-Y., Yang, H.-M., Xu, H., Xue, F., Li, S., & Feng, X.-H. (2014). Gene cloning, expression, and characterization of an exo-inulinase from Paenibacillus polymyxa ZJ-9. Applied Biochemistry and Biotechnology, 173(6), 1419–1430.
Mancilla-Margalli, N. A., & López, M. G. (2006). Water-soluble carbohydrates and fructan structure patterns from Agave and Dasylirion species. Journal of Agricultural and Food Chemistry, 54(20), 7832–7839.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry, 31(3), 426–428.
Chen, C.-C., Hwang, J.-K., & Yang, J.-M. (2006). (PS)2: protein structure prediction server. Nucleic Acids Research, 34(Suppl 2), W152–W157.
Chen, V. B., Arendall, W. B., III, Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S., & Richardson, D. C. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D: Structural Biology, 66(1), 12–21.
Hess, B., Kutzner, C., van der Spoel, D., & Lindahl, E. (2008). GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation, 4(3), 435–447.
MacKerell, A. D., Jr., Feig, M., & Brooks, C. L., III. (2004). Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. Journal of Computational Chemistry, 25(11), 1400–1415.
Hess, B. (2008). P-LINCS: a parallel linear constraint solver for molecular simulation. Journal of Chemical Theory and Computation, 4(1), 116–122.
Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593.
Thomsen, R., & Christensen, M. H. (2006). MolDock: a new technique for high-accuracy molecular docking. Journal of Medicinal Chemistry, 49(11), 3315–3321.
Yang, J.-M., & Chen, C.-C. (2004). GEMDOCK: a generic evolutionary method for molecular docking. Proteins: Structure, Function, and Bioinformatics, 55(2), 288–304.
Wavefunction, Inc. Spartan’08. Available from www.wavefun.com. Accessed 30 Dec 2015.
Stephens, P. J., Devlin, F. J., Chabalowski, C. F., & Frisch, M. J. (1994). Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. The Journal of Physical Chemistry, 98(45), 11623–11627.
Petersson, G. A., Tensfeldt, T. G., & Montgomery, J. A., Jr. (1991). A complete basis set model chemistry. III. The complete basis set‐quadratic configuration interaction family of methods. The Journal of Chemical Physics, 94(9), 6091–6101.
Omori, T., Ueno, K., Muramatsu, K., Kikuchi, M., Onodera, S., & Shiomi, N. (2010). Characterization of recombinant β-fructofuranosidase from Bifidobacterium adolescentis G1. Chemistry Central Journal, 4(1), 9.
Saulnier, D. M. A., Molenaar, D., de Vos, W. M., Gibson, G. R., & Kolida, S. (2007). Identification of prebiotic fructooligosaccharide metabolism in Lactobacillus plantarum WCFS1 through microarrays. Applied and Environmental Microbiology, 73(6), 1753–1765.
McNicholas, S., Potterton, E., Wilson, K. S., & Noble, M. E. M. (2011). Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallographica Section D: Structural Biology, 67(4), 386–394.
Nagem, R. A. P., Rojas, A. L., Golubev, A. M., Korneeva, O. S., Eneyskaya, E. V., Kulminskaya, A. A., Neustroev, K. K., & Polikarpov, I. (2004). Crystal structure of exo-inulinase from Aspergillus awamori: the enzyme fold and structural determinants of substrate recognition. Journal of Molecular Biology, 344(2), 471–480.
Chaudhuri, I., Söding, J., & Lupas, A. N. (2008). Evolution of the β-propeller fold. Proteins: Structure, Function, and Bioinformatics, 71(2), 795–803.
Chen, C. K.-M., Chan, N.-L., & Wang, A. H.-J. (2011). The many blades of the β-propeller proteins: conserved but versatile. Trends in Biochemical Sciences, 36(10), 553–561.
Cuskin, F., Flint, J. E., Gloster, T. M., Morland, C., Baslé, A., Henrissat, B., Coutinho, P. M., Strazzulli, A., Solovyova, A. S., Davies, G. J., & Gilbert, H. J. (2012). How nature can exploit nonspecific catalytic and carbohydrate binding modules to create enzymatic specificity. Proceedings of the National Academy of Sciences USA, 109(51), 20889–20894.
Sainz-Polo, M. A., Ramírez-Escudero, M., Lafraya, A., González, B., Marín-Navarro, J., Polaina, J., & Sanz-Aparicio, J. (2013). Three-dimensional structure of Saccharomyces invertase: role of a non-catalytic domain in oligomerization and substrate specificity. Journal of Biological Chemistry, 288(14), 9755–9766.
Privalov, P. L. 1979. in Stability of proteins small globular proteins, vol 33: advances in protein chemistry (Anfinsen, C. B., Edsall, J. T., & Richards, F. M. eds.), Academic Press, New York, pp. 167–241.
Ferré-D’Amaré, A. R., & Burley, S. K. (1994). Use of dynamic light scattering to assess crystallizability of macromolecules and macromolecular assemblies. Structure, 2(5), 357–359.
Talley, K., & Alexov, E. (2010). On the pH-optimum of activity and stability of proteins. Proteins: Structure, Function, and Bioinformatics, 78(12), 2699–2706.
Aziz, S., Jalal, F., Nawaz, M., Niaz, B., Shah, F.A., Memon, M. H.-u.-R., Latif, F., Nadeem, S., & Rajoka, M.I. (2011). Hyperproduction and thermal characterization of a novel invertase from a double mutant derivative of Kluyveromyces marxianus. Food Technology and Biotechnology, 49(4), 465–473.
Hussain, A., Rashid, M. H., Perveen, R., & Ashraf, M. (2009). Purification, kinetic and thermodynamic characterization of soluble acid invertase from sugarcane (Saccharum officinarum L.). Plant Physiology and Biochemistry, 47(3), 188–194.
Rodríguez-Chong, A., Ramírez, J. A., Garrote, G., & Vázquez, M. (2004). Hydrolysis of sugar cane bagasse using nitric acid: a kinetic assessment. Journal of Food Engineering, 61(2), 143–152.
Tombari, E., Salvetti, G., Ferrari, C., & Johari, G. P. (2007). Kinetics and thermodynamics of sucrose hydrolysis from real-time enthalpy and heat capacity measurements. The Journal of Physical Chemistry B, 111(3), 496–501.
Garcia-Viloca, M., Gao, J., Karplus, M., & Truhlar, D. G. (2004). How enzymes work: analysis by modern rate theory and computer simulations. Science, 303(5655), 186–195.
Carson, M., Johnson, D. H., McDonald, H., Brouillette, C., & DeLucas, L. J. (2007). His-tag impact on structure. Acta Crystallographica Section D: Structural Biology, 63(3), 295–301.
Mason, A. B., He, Q.-Y., Halbrooks, P. J., Everse, S. J., Gumerov, D. R., Kaltashov, I. A., Smith, V. C., Hewitt, J., & MacGillivray, R. T. A. (2002). Differential effect of a His tag at the N- and C-termini: functional studies with recombinant human serum transferrin. Biochemistry, 41(3), 9448–9454.
Sabaty, M., Grosse, S., Adryanczyk, G., Boiry, S., Biaso, F., Arnoux, P., & Pignol, D. (2013). Detrimental effect of the 6 His C-terminal tag on YedY enzymatic activity and influence of the TAT signal sequence on YedY synthesis. BMC Biochemistry, 14(1), 28.
Xu, C.-G., Fan, X.-J., Fu, Y.-J., & Liang, A.-H. (2008). Effect of location of the His-tag on the production of soluble and functional Buthus martensii Karsch insect toxin. Protein Expression and Purification, 59(1), 103–109.
Bujacz, A., Jedrzejczak-Krzepkowska, M., Bielecki, S., Redzynia, I., & Bujacz, G. (2011). Crystal structures of the apo form of β-fructofuranosidase from Bifidobacterium longum and its complex with fructose. The FEBS Journal, 278(10), 1728–1744.
Fülöp, V., Böcskei, Z., & Polgár, L. (1998). Prolyl oligopeptidase: an unusual β-propeller domain regulates proteolysis. Cell, 94(2), 161–170.
Acknowledgments
This work was funded by FRABA 14-2013 grant of University of Colima, Mexico, and by CONACyT PhD scholarship (registration number 270848). We thank Dr. Jean Jakoncic (BNL, NSLS-II) for his support on X-ray data collection using synchrotron radiation in order to go further on the crystallographic work for this investigation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Mendoza-Llerenas, E.O., Pérez, D.J., Gómez-Sandoval, Z. et al. Lactobacillus plantarum WCFS1 β-Fructosidase: Evidence for an Open Funnel-Like Channel Through the Catalytic Domain with Importance for the Substrate Selectivity. Appl Biochem Biotechnol 180, 1056–1075 (2016). https://doi.org/10.1007/s12010-016-2152-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2152-2